Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria
- PMID: 16820502
- PMCID: PMC1489352
- DOI: 10.1128/AEM.00682-06
Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria
Abstract
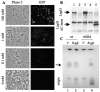
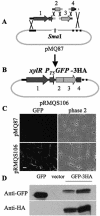
A tool kit of vectors was designed to manipulate and express genes from a wide range of gram-negative species by using in vivo recombination. Saccharomyces cerevisiae can use its native recombination proteins to combine several amplicons in a single transformation step with high efficiency. We show that this technology is particularly useful for vector design. Shuttle, suicide, and expression vectors useful in a diverse group of bacteria are described and utilized. This report describes the use of these vectors to mutate clpX and clpP of the opportunistic pathogen Pseudomonas aeruginosa and to explore their roles in biofilm formation and surface motility. Complementation of the rhamnolipid biosynthetic gene rhlB is also described. Expression vectors are used for controlled expression of genes in two pseudomonad species. To demonstrate the facility of building complicated constructs with this technique, the recombination of four PCR-generated amplicons in a single step at >80% efficiency into one of these vectors is shown. These tools can be used for genetic studies of pseudomonads and many other gram-negative bacteria.
Figures


References
-
- Bascom-Slack, C. A., and D. Dawson. 1998. A physical assay for detection of early meiotic recombination intermediates in Saccharomyces cerevisiae. Mol. Gen. Genet. 258:512-520. - PubMed
-
- Burke, D., D. Dawson, and T. Stearns. 2000. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
-
- Choi, K. H., A. Kumar, and H. P. Schweizer. 2005. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64:391-397. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

