Purine biosynthesis, riboflavin production, and trophic-phase span are controlled by a Myb-related transcription factor in the fungus Ashbya gossypii
- PMID: 16820505
- PMCID: PMC1489300
- DOI: 10.1128/AEM.00424-06
Purine biosynthesis, riboflavin production, and trophic-phase span are controlled by a Myb-related transcription factor in the fungus Ashbya gossypii
Abstract
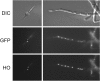
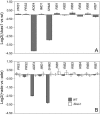
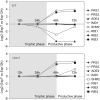
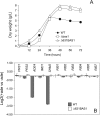
Ashbya gossypii is a natural riboflavin overproducer used in the industrial production of the vitamin. We have isolated an insertional mutant exhibiting higher levels of riboflavin production than the wild type. DNA analysis of the targeted locus in the mutant strain revealed that a syntenic homolog of the Saccharomyces cerevisiae BAS1 gene, a member of the Myb family of transcription factors, was inactivated. Directed gene disruption of AgBAS1 confirmed the phenotype observed for the insertional mutant, and the Deltabas1 mutant also showed auxotrophy for adenine and several growth defects, such as a delay in the germination of the spores and an abnormally prolonged trophic phase. Additionally, we demonstrate that the DNA-binding domain of AgBas1p is able to bind to the Bas1-binding motifs in the AgADE4 promoter; we also show a clear nuclear localization of a green fluorescent protein-Bas1 fusion protein. Real-time quantitative PCR analyses comparing the wild type and the Deltabas1 mutant revealed that AgBAS1 was responsible for the adenine-mediated regulation of the purine and glycine pathways, since the transcription of the ADE4 and SHM2 genes was virtually abolished in the Deltabas1 mutant. Furthermore, the transcription of ADE4 and SHM2 in the Deltabas1 mutant did not diminish during the transition from the trophic to the productive phase did not diminish, in contrast to what occurred in the wild-type strain. A C-terminal deletion in the AgBAS1 gene, comprising a hypothetical regulatory domain, caused constitutive activation of the purine and glycine pathways, enhanced riboflavin overproduction, and prolonged the trophic phase. Taking these results together, we propose that in A. gossypii, AgBAS1 is an important transcription factor that is involved in the regulation of different physiological processes, such as purine and glycine biosynthesis, riboflavin overproduction, and growth.
Figures



Similar articles
-
Metabolic engineering of the purine pathway for riboflavin production in Ashbya gossypii.Appl Environ Microbiol. 2005 Oct;71(10):5743-51. doi: 10.1128/AEM.71.10.5743-5751.2005. Appl Environ Microbiol. 2005. PMID: 16204483 Free PMC article.
-
Growth stress triggers riboflavin overproduction in Ashbya gossypii.Appl Microbiol Biotechnol. 2007 Sep;76(3):569-78. doi: 10.1007/s00253-007-1075-9. Epub 2007 Jul 18. Appl Microbiol Biotechnol. 2007. PMID: 17639374
-
The improvement of riboflavin production in Ashbya gossypii via disparity mutagenesis and DNA microarray analysis.Appl Microbiol Biotechnol. 2011 Sep;91(5):1315-26. doi: 10.1007/s00253-011-3325-0. Epub 2011 May 15. Appl Microbiol Biotechnol. 2011. PMID: 21573938
-
Three biotechnical processes using Ashbya gossypii, Candida famata, or Bacillus subtilis compete with chemical riboflavin production.Appl Microbiol Biotechnol. 2000 May;53(5):509-16. doi: 10.1007/s002530051649. Appl Microbiol Biotechnol. 2000. PMID: 10855708 Review.
-
Ashbya gossypii: a model for fungal developmental biology.Nat Rev Microbiol. 2005 May;3(5):421-9. doi: 10.1038/nrmicro1148. Nat Rev Microbiol. 2005. PMID: 15821727 Review.
Cited by
-
Utilization of xylose by engineered strains of Ashbya gossypii for the production of microbial oils.Biotechnol Biofuels. 2017 Jan 3;10:3. doi: 10.1186/s13068-016-0685-9. eCollection 2017. Biotechnol Biofuels. 2017. PMID: 28053663 Free PMC article.
-
Metabolic Engineering of Bacillus subtilis for Riboflavin Production: A Review.Microorganisms. 2023 Jan 8;11(1):164. doi: 10.3390/microorganisms11010164. Microorganisms. 2023. PMID: 36677456 Free PMC article. Review.
-
Genome evolution in the eremothecium clade of the Saccharomyces complex revealed by comparative genomics.G3 (Bethesda). 2011 Dec;1(7):539-48. doi: 10.1534/g3.111.001032. Epub 2011 Dec 1. G3 (Bethesda). 2011. PMID: 22384365 Free PMC article.
-
Metabolic engineering of Ashbya gossypii for limonene production from xylose.Biotechnol Biofuels Bioprod. 2022 Jul 15;15(1):79. doi: 10.1186/s13068-022-02176-0. Biotechnol Biofuels Bioprod. 2022. PMID: 35841062 Free PMC article.
-
Genomic analysis of a riboflavin-overproducing Ashbya gossypii mutant isolated by disparity mutagenesis.BMC Genomics. 2020 Apr 23;21(1):319. doi: 10.1186/s12864-020-6709-7. BMC Genomics. 2020. PMID: 32326906 Free PMC article.
References
-
- Allison, A. C., and E. M. Eugui. 2000. Mycophenolate mofetil and its mechanisms of action. Immunopharmacology 47:85-118. - PubMed
-
- Arndt, K. T., C. Styles, and G. R. Fink. 1987. Multiple global regulators control HIS4 transcription in yeast. Science 237:874-880. - PubMed
-
- Bacher, A., S. Eberhardt, M. Fischer, K. Kis, and G. Richter. 2000. Biosynthesis of vitamin B2 (riboflavin). Annu. Rev. Nutr. 20:153-167. - PubMed
-
- Demain, A. L. 1972. Riboflavin oversynthesis. Annu. Rev. Microbiol. 26:369-388. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

