Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB
- PMID: 16820507
- PMCID: PMC1489311
- DOI: 10.1128/AEM.03006-05
Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB
Abstract
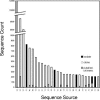
A 16S rRNA gene database (http://greengenes.lbl.gov) addresses limitations of public repositories by providing chimera screening, standard alignment, and taxonomic classification using multiple published taxonomies. It was found that there is incongruent taxonomic nomenclature among curators even at the phylum level. Putative chimeras were identified in 3% of environmental sequences and in 0.2% of records derived from isolates. Environmental sequences were classified into 100 phylum-level lineages in the Archaea and Bacteria.
Figures
References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Brodie, E., S. Edwards, and N. Clipson. 2002. Bacterial community dynamics across a floristic gradient in a temperate upland grassland ecosystem. Microb. Ecol. 44:260-270. - PubMed
-
- Castiglioni, B., E. Rizzi, A. Frosini, K. Sivonen, P. Rajaniemi, A. Rantala, M. A. Mugnai, S. Ventura, A. Wilmotte, C. Boutte, S. Grubisic, P. Balthasart, C. Consolandi, R. Bordoni, A. Mezzelani, C. Battaglia, and G. De Bellis. 2004. Development of a universal microarray based on the ligation detection reaction and 16S rRNA gene polymorphism to target diversity of cyanobacteria. Appl. Environ. Microbiol. 70:7161-7172. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

