SAP30L interacts with members of the Sin3A corepressor complex and targets Sin3A to the nucleolus
- PMID: 16820529
- PMCID: PMC1500868
- DOI: 10.1093/nar/gkl401
SAP30L interacts with members of the Sin3A corepressor complex and targets Sin3A to the nucleolus
Abstract
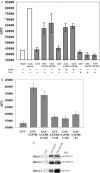
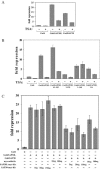
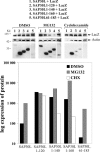
Histone acetylation plays a key role in the regulation of gene expression. The chromatin structure and accessibility of genes to transcription factors is regulated by enzymes that acetylate and deacetylate histones. The Sin3A corepressor complex recruits histone deacetylases and in many cases represses transcription. Here, we report that SAP30L, a close homolog of Sin3-associated protein 30 (SAP30), interacts with several components of the Sin3A corepressor complex. We show that it binds to the PAH3/HID (Paired Amphipathic Helix 3/Histone deacetylase Interacting Domain) region of mouse Sin3A with residues 120-140 in the C-terminal part of the protein. We provide evidence that SAP30L induces transcriptional repression, possibly via recruitment of Sin3A and histone deacetylases. Finally, we characterize a functional nucleolar localization signal in SAP30L and show that SAP30L and SAP30 are able to target Sin3A to the nucleolus.
Figures




Similar articles
-
Alternative mRNA splicing of SAP30L regulates its transcriptional repression activity.FEBS Lett. 2008 Jan 23;582(2):379-84. doi: 10.1016/j.febslet.2007.11.084. Epub 2007 Dec 10. FEBS Lett. 2008. PMID: 18070604
-
Individual subunits of the Ssn6-Tup11/12 corepressor are selectively required for repression of different target genes.Mol Cell Biol. 2007 Feb;27(3):1069-82. doi: 10.1128/MCB.01674-06. Epub 2006 Nov 13. Mol Cell Biol. 2007. PMID: 17101775 Free PMC article.
-
The homeobox gene Mohawk represses transcription by recruiting the sin3A/HDAC co-repressor complex.Dev Dyn. 2009 Mar;238(3):572-80. doi: 10.1002/dvdy.21873. Dev Dyn. 2009. PMID: 19235719
-
The coregulator Alien.Nucl Recept Signal. 2007 Nov 30;5:e008. doi: 10.1621/nrs.05008. Nucl Recept Signal. 2007. PMID: 18174916 Free PMC article. Review.
-
Sin3: insight into its transcription regulatory functions.Eur J Cell Biol. 2013 Aug-Sep;92(8-9):237-46. doi: 10.1016/j.ejcb.2013.09.001. Epub 2013 Oct 9. Eur J Cell Biol. 2013. PMID: 24189169 Review.
Cited by
-
The type 1 diabetes - HLA susceptibility interactome--identification of HLA genotype-specific disease genes for type 1 diabetes.PLoS One. 2010 Mar 5;5(3):e9576. doi: 10.1371/journal.pone.0009576. PLoS One. 2010. PMID: 20221424 Free PMC article.
-
DNA-binding and -bending activities of SAP30L and SAP30 are mediated by a zinc-dependent module and monophosphoinositides.Mol Cell Biol. 2009 Jan;29(2):342-56. doi: 10.1128/MCB.01213-08. Epub 2008 Nov 17. Mol Cell Biol. 2009. PMID: 19015240 Free PMC article.
-
Sin3: master scaffold and transcriptional corepressor.Biochim Biophys Acta. 2009 Jun-Aug;1789(6-8):443-50. doi: 10.1016/j.bbagrm.2009.05.007. Epub 2009 Jun 6. Biochim Biophys Acta. 2009. PMID: 19505602 Free PMC article. Review.
-
Unconventional metabolites in chromatin regulation.Biosci Rep. 2022 Jan 28;42(1):BSR20211558. doi: 10.1042/BSR20211558. Biosci Rep. 2022. PMID: 34988581 Free PMC article.
-
Inositol phosphates and core subunits of the Sin3L/Rpd3L histone deacetylase (HDAC) complex up-regulate deacetylase activity.J Biol Chem. 2019 Sep 20;294(38):13928-13938. doi: 10.1074/jbc.RA119.009780. Epub 2019 Jul 29. J Biol Chem. 2019. PMID: 31358618 Free PMC article.
References
-
- Ayer D.E. Histone deacetylases: transcriptional repression with SINers and NuRDs. Trends. Cell Biol. 1999;9:193–198. - PubMed
-
- Narlikar G.J., Fan H.Y., Kingston R.E. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–487. - PubMed
-
- Silverstein R.A., Ekwall K. Sin3: a flexible regulator of global gene expression and genome stability. Curr. Genet. 2005;47:1–17. - PubMed
-
- Zhang Y., Iratni R., Erdjument-Bromage H., Tempst P., Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–364. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

