Structure of the synthetase domain of human CTP synthetase, a target for anticancer therapy
- PMID: 16820675
- PMCID: PMC2242944
- DOI: 10.1107/S1744309106018136
Structure of the synthetase domain of human CTP synthetase, a target for anticancer therapy
Abstract
Cytidine triphosphate synthetase (CTPS) is a key enzyme in nucleic acid and phospholipid biosynthesis and its activity is increased in certain human cancers, making it a promising drug target. The crystal structure of the synthetase domain of human CTPS, which represents the first structure of a CTPS from an eukaryote, has been determined. The structure is homotetrameric and each active site is formed by three different subunits. Sulfate ions bound to the active sites indicate the positions of phosphate-binding sites for the substrates ATP and UTP and the feedback inhibitor CTP. Together with earlier structures of bacterial CTPS, the human CTPS structure provides an extended understanding of the structure-function relationship of CTPS-family members. The structure also serves as a basis for structure-based design of anti-proliferative inhibitors.
Figures
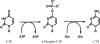
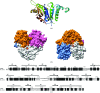
Similar articles
-
Crystal structures of CTP synthetase reveal ATP, UTP, and glutamine binding sites.Structure. 2004 Aug;12(8):1413-23. doi: 10.1016/j.str.2004.05.013. Structure. 2004. PMID: 15296735
-
Crystal structure of Escherichia coli cytidine triphosphate synthetase, a nucleotide-regulated glutamine amidotransferase/ATP-dependent amidoligase fusion protein and homologue of anticancer and antiparasitic drug targets.Biochemistry. 2004 Jun 1;43(21):6447-63. doi: 10.1021/bi0496945. Biochemistry. 2004. PMID: 15157079 Free PMC article.
-
The role of lysine residues 297 and 306 in nucleoside triphosphate regulation of E. coli CTP synthase: inactivation by 2',3'-dialdehyde ATP and mutational analyses.Biochim Biophys Acta. 2006 Feb;1764(2):199-210. doi: 10.1016/j.bbapap.2005.11.021. Epub 2005 Dec 27. Biochim Biophys Acta. 2006. PMID: 16427816
-
CTPS cytoophidia in Drosophila: distribution, regulation, and physiological roles.Exp Cell Res. 2025 Apr 15;447(2):114536. doi: 10.1016/j.yexcr.2025.114536. Epub 2025 Mar 22. Exp Cell Res. 2025. PMID: 40122502 Review.
-
Cyclopentenyl cytosine (CPEC): an overview of its in vitro and in vivo activity.Curr Cancer Drug Targets. 2007 Aug;7(5):504-9. doi: 10.2174/156800907781386579. Curr Cancer Drug Targets. 2007. PMID: 17691910 Review.
Cited by
-
Molecular Classification of Primary Immunodeficiencies of T Lymphocytes.Adv Immunol. 2018;138:99-193. doi: 10.1016/bs.ai.2018.02.003. Epub 2018 Mar 29. Adv Immunol. 2018. PMID: 29731008 Free PMC article. Review.
-
CTP synthase 1 deficiency in humans reveals its central role in lymphocyte proliferation.Nature. 2014 Jun 12;510(7504):288-92. doi: 10.1038/nature13386. Epub 2014 May 28. Nature. 2014. PMID: 24870241 Free PMC article.
-
Phosphorylation of human CTP synthetase 1 by protein kinase A: identification of Thr455 as a major site of phosphorylation.J Biol Chem. 2007 Feb 23;282(8):5367-77. doi: 10.1074/jbc.M610993200. Epub 2006 Dec 22. J Biol Chem. 2007. PMID: 17189248 Free PMC article.
-
Phosphorylation of human CTP synthetase 1 by protein kinase C: identification of Ser(462) and Thr(455) as major sites of phosphorylation.J Biol Chem. 2007 Jun 15;282(24):17613-22. doi: 10.1074/jbc.M702799200. Epub 2007 Apr 26. J Biol Chem. 2007. PMID: 17463002 Free PMC article.
-
Filamentation of hCTPS1 with CTP.Cell Biosci. 2025 Jul 30;15(1):112. doi: 10.1186/s13578-025-01450-6. Cell Biosci. 2025. PMID: 40739251 Free PMC article.
References
-
- Brünger, A. T. (1992). Nature (London), 355, 472–474. - PubMed
-
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. - PubMed
-
- Dereuddre-Bosquet, N., Roy, B., Routledge, K., Clayette, P., Foucault, G. & Lepoivre, M. (2004). Antiviral Res.61, 67–70. - PubMed
-
- Diederichs, K. & Karplus, P. A. (1997). Nature Struct. Biol.4, 269–275. - PubMed
-
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

