Structure of human MIP-3alpha chemokine
- PMID: 16820679
- PMCID: PMC2242953
- DOI: 10.1107/S1744309106006890
Structure of human MIP-3alpha chemokine
Abstract
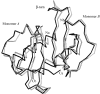
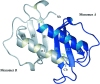
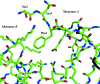
The structure of the human macrophage inflammatory protein-3alpha (MIP-3alpha) has been determined at 1.81 angstroms resolution by X-ray crystallography. The dimer crystallized in the tetragonal space group I4, with unit-cell parameters a = b = 83.99, c = 57.20 angstroms. The crystals exhibit two molecules in the asymmetric unit. The structure was solved by the molecular-replacement method and the model was refined to a conventional R value of 20.6% (R(free) = 25.7%). MIP-3alpha possesses the same monomeric structure as previously described for other chemokines. However, in addition to limited structural changes in the beta1-beta2 hairpin of monomer B, the electron density is fully defined for a few extra residues at the N- and C-termini of monomer A and the C-terminus of monomer B compared with MIP-3alpha in space group P6(1). As the N-terminal and loop regions have been shown to be critical for receptor binding and signaling, this additional structural information may help in determining the basis of the CCR6 selectivity of MIP-3alpha.
Figures
Similar articles
-
NMR solution structure of murine CCL20/MIP-3alpha, a chemokine that specifically chemoattracts immature dendritic cells and lymphocytes through its highly specific interaction with the beta-chemokine receptor CCR6.J Biol Chem. 2001 Jul 27;276(30):28372-9. doi: 10.1074/jbc.M103121200. Epub 2001 May 23. J Biol Chem. 2001. PMID: 11373289
-
The structure of human macrophage inflammatory protein-3alpha /CCL20. Linking antimicrobial and CC chemokine receptor-6-binding activities with human beta-defensins.J Biol Chem. 2002 Oct 4;277(40):37647-54. doi: 10.1074/jbc.M203907200. Epub 2002 Jul 30. J Biol Chem. 2002. PMID: 12149255
-
Chemokine expression in human erythroid leukemia cell line AS-E2: macrophage inflammatory protein-3alpha/CCL20 is induced by inflammatory cytokines.Exp Hematol. 2006 Jan;34(1):19-26. doi: 10.1016/j.exphem.2005.09.012. Exp Hematol. 2006. PMID: 16413387
-
The CC chemokine CCL20 and its receptor CCR6.Cytokine Growth Factor Rev. 2003 Oct;14(5):409-26. doi: 10.1016/s1359-6101(03)00049-2. Cytokine Growth Factor Rev. 2003. PMID: 12948524 Review.
-
The airway epithelium as immune modulator: the LARC ascending.Am J Respir Cell Mol Biol. 2003 Jun;28(6):641-4. doi: 10.1165/rcmb.F271. Am J Respir Cell Mol Biol. 2003. PMID: 12760960 Review. No abstract available.
Cited by
-
Prognostic Value of Macrophage Inflammatory Protein-3alpha (MIP3-Alpha) and Severity Scores in Elderly Patients with Sepsis.J Inflamm Res. 2024 Mar 8;17:1503-1509. doi: 10.2147/JIR.S447142. eCollection 2024. J Inflamm Res. 2024. PMID: 38476471 Free PMC article.
-
Protein engineering of the chemokine CCL20 prevents psoriasiform dermatitis in an IL-23-dependent murine model.Proc Natl Acad Sci U S A. 2017 Nov 21;114(47):12460-12465. doi: 10.1073/pnas.1704958114. Epub 2017 Nov 6. Proc Natl Acad Sci U S A. 2017. PMID: 29109267 Free PMC article.
-
The Chemokine, CCL20, and Its Receptor, CCR6, in the Pathogenesis and Treatment of Psoriasis and Psoriatic Arthritis.J Psoriasis Psoriatic Arthritis. 2023 Jul;8(3):107-117. doi: 10.1177/24755303231159106. Epub 2023 Mar 12. J Psoriasis Psoriatic Arthritis. 2023. PMID: 39296310 Free PMC article. Review.
-
Exploiting agonist biased signaling of chemokines to target cancer.Mol Carcinog. 2017 Mar;56(3):804-813. doi: 10.1002/mc.22571. Epub 2016 Oct 4. Mol Carcinog. 2017. PMID: 27648825 Free PMC article.
-
Heterodimers Are an Integral Component of Chemokine Signaling Repertoire.Int J Mol Sci. 2023 Jul 19;24(14):11639. doi: 10.3390/ijms241411639. Int J Mol Sci. 2023. PMID: 37511398 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

