Exploring the regulation of human neural precursor cell differentiation using arrays of signaling microenvironments
- PMID: 16820778
- PMCID: PMC1681508
- DOI: 10.1038/msb4100076
Exploring the regulation of human neural precursor cell differentiation using arrays of signaling microenvironments
Abstract
Cells of a developing embryo integrate a complex array of local and long-range signals that act in concert with cell-intrinsic determinants to influence developmental decisions. To systematically investigate the effects of molecular microenvironments on cell fate decisions, we developed an experimental method based on parallel exposure of cells to diverse combinations of extracellular signals followed by quantitative, multi-parameter analysis of cellular responses. Primary human neural precursor cells were captured and cultured on printed microenvironment arrays composed of mixtures of extracellular matrix components, morphogens, and other signaling proteins. Quantitative single cell analysis revealed striking effects of some of these signals on the extent and direction of differentiation. We found that Wnt and Notch co-stimulation could maintain the cells in an undifferentiated-like, proliferative state, whereas bone morphogenetic protein 4 induced an 'indeterminate' differentiation phenotype characterized by simultaneous expression of glial and neuronal markers. Multi-parameter analysis of responses to conflicting signals revealed interactions more complex than previously envisaged including dominance relations that may reflect a cell-intrinsic system for robust specification of responses in complex microenvironments.
Figures
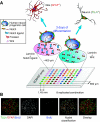
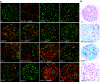
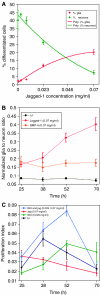
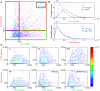


Comment in
-
Microarraying the cellular microenvironment.Mol Syst Biol. 2006;2:39. doi: 10.1038/msb4100079. Epub 2006 Jul 4. Mol Syst Biol. 2006. PMID: 16820780 Free PMC article. No abstract available.
References
-
- Anderson DG, Levenberg S, Langer R (2004) Nanoliter-scale synthesis of arrayed biomaterials and application to human embryonic stem cells. Nat Biotechnol 22: 863–866 - PubMed
-
- Anderson DJ (2001) Stem cells and pattern formation in the nervous system: the possible versus the actual. Neuron 30: 19–35 - PubMed
-
- Bertrand N, Castro DS, Guillemont F (2002) Proneural genes and the specification of neuronal cell types. Nat Rev Neurosci 3: 517–530 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

