Origin of mitochondria by intracellular enslavement of a photosynthetic purple bacterium
- PMID: 16822756
- PMCID: PMC1634775
- DOI: 10.1098/rspb.2006.3531
Origin of mitochondria by intracellular enslavement of a photosynthetic purple bacterium
Abstract
Mitochondria originated by permanent enslavement of purple non-sulphur bacteria. These endosymbionts became organelles through the origin of complex protein-import machinery and insertion into their inner membranes of protein carriers for extracting energy for the host. A chicken-and-egg problem exists: selective advantages for evolving import machinery were absent until inner membrane carriers were present, but this very machinery is now required for carrier insertion. I argue here that this problem was probably circumvented by conversion of the symbiont protein-export machinery into protein-import machinery, in three phases. I suggest that the first carrier entered the periplasmic space via pre-existing beta-barrel proteins in the bacterial outer membrane that later became Tom40, and inserted into the inner membrane probably helped by a pre-existing inner membrane protein, thereby immediately providing the protoeukaryote host with photosynthesate. This would have created a powerful selective advantage for evolving more efficient carrier import by inserting Tom70 receptors. Massive gene transfer to the nucleus inevitably occurred by mutation pressure. Finally, pressure from harmful, non-selected gene transfer to the nucleus probably caused evolution of the presequence mechanism, and photosynthesis was lost.
Figures
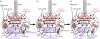
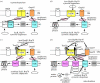
Similar articles
-
Mitochondrial Machineries for Protein Import and Assembly.Annu Rev Biochem. 2017 Jun 20;86:685-714. doi: 10.1146/annurev-biochem-060815-014352. Epub 2017 Mar 15. Annu Rev Biochem. 2017. PMID: 28301740 Review.
-
A novel insertion pathway of mitochondrial outer membrane proteins with multiple transmembrane segments.J Cell Biol. 2007 Dec 31;179(7):1355-63. doi: 10.1083/jcb.200702143. Epub 2007 Dec 24. J Cell Biol. 2007. PMID: 18158327 Free PMC article.
-
Biogenesis of the mitochondrial TOM complex: Mim1 promotes insertion and assembly of signal-anchored receptors.J Biol Chem. 2008 Jan 4;283(1):120-127. doi: 10.1074/jbc.M706997200. Epub 2007 Nov 1. J Biol Chem. 2008. PMID: 17974559
-
Evolution of mitochondrial protein biogenesis.Biochim Biophys Acta. 2009 Jun;1790(6):409-15. doi: 10.1016/j.bbagen.2009.04.004. Epub 2009 Apr 10. Biochim Biophys Acta. 2009. PMID: 19362582 Review.
-
Yeast mitochondria can process de novo designed β-barrel proteins.FEBS J. 2024 Jan;291(2):292-307. doi: 10.1111/febs.16950. Epub 2023 Oct 2. FEBS J. 2024. PMID: 37723586
Cited by
-
Deep phylogeny, ancestral groups and the four ages of life.Philos Trans R Soc Lond B Biol Sci. 2010 Jan 12;365(1537):111-32. doi: 10.1098/rstb.2009.0161. Philos Trans R Soc Lond B Biol Sci. 2010. PMID: 20008390 Free PMC article.
-
Cell evolution and Earth history: stasis and revolution.Philos Trans R Soc Lond B Biol Sci. 2006 Jun 29;361(1470):969-1006. doi: 10.1098/rstb.2006.1842. Philos Trans R Soc Lond B Biol Sci. 2006. PMID: 16754610 Free PMC article. Review.
-
LACTB is a filament-forming protein localized in mitochondria.Proc Natl Acad Sci U S A. 2009 Nov 10;106(45):18960-5. doi: 10.1073/pnas.0906734106. Epub 2009 Oct 26. Proc Natl Acad Sci U S A. 2009. PMID: 19858488 Free PMC article.
-
Near infrared radiation rescues mitochondrial dysfunction in cortical neurons after oxygen-glucose deprivation.Metab Brain Dis. 2015 Apr;30(2):491-6. doi: 10.1007/s11011-014-9515-6. Epub 2014 Mar 6. Metab Brain Dis. 2015. PMID: 24599760 Free PMC article.
-
Critical Role of Mitochondrial Fatty Acid Metabolism in Normal Cell Function and Pathological Conditions.Int J Mol Sci. 2024 Jun 12;25(12):6498. doi: 10.3390/ijms25126498. Int J Mol Sci. 2024. PMID: 38928204 Free PMC article. Review.
References
-
- Aaronson S, Dhawale S.W, Patni N.J, DeAngelis B, Frank O, Baker H. The cell content and secretion of water-soluble vitamins by several freshwater algae. Arch. Microbiol. 1977;112:57–59. 10.1007/BF00446654 - DOI - PubMed
-
- Adams K.L, Palmer J.D. Evolution of mitochondrial gene content: gene loss and transfer to the nucleus. Mol. Phylogenet. Evol. 2003;29:380–395. 10.1016/S1055-7903(03)00194-5 - DOI - PubMed
-
- Ahting U, Waizenegger T, Neupert W, Rapaport D. Signal-anchored proteins follow a unique insertion pathway into the outer membrane of mitochondria. J. Biol. Chem. 2005;280:48–53. - PubMed
-
- Allen J.F, Puthiyaveetil S, Strom J, Allen C.A. Energy transduction anchors genes in organelles. BioEssays. 2005;27:426–435. 10.1002/bies.20194 - DOI - PubMed
-
- Andersson S.G, Karlberg O, Canback B, Kurland C.G. On the origin of mitochondria: a genomics perspective. Phil. Trans. R. Soc. B. 2003;358:165–177. 10.1098/rstb.2002.1193 (See discussion on pages 177–179.) - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

