Structure of the angiogenesis inhibitor ovalicin bound to its noncognate target, human Type 1 methionine aminopeptidase
- PMID: 16823043
- PMCID: PMC2242583
- DOI: 10.1110/ps.062278006
Structure of the angiogenesis inhibitor ovalicin bound to its noncognate target, human Type 1 methionine aminopeptidase
Abstract
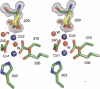
Methionine aminopeptidases (MetAPs) remove the initiator methionine during protein biosynthesis. They exist in two isoforms, MetAP1 and MetAP2. The anti-angiogenic compound fumagillin binds tightly to the Type 2 MetAPs but only weakly to Type 1. High-affinity complexes of fumagillin and its relative ovalicin with Type 2 human MetAP have been reported. Here we describe the crystallographic structure of the low-affinity complex between ovalicin and Type 1 human MetAP at 1.1 A resolution. This provides the first opportunity to compare the structures of ovalicin or fumagillin bound to a Type 1 and a Type 2 MetAP. For both Type 1 and Type 2 human MetAPs the inhibitor makes a covalent adduct with a corresponding histidine. At the same time there are significant differences in the alignment of the inhibitors within the respective active sites. It has been argued that the lower affinity of ovalicin and fumagillin for the Type 1 MetAPs is due to the smaller size of their active sites relative to the Type 2 enzymes. Comparison with the uncomplexed structure of human Type 1 MetAP indicates that there is some truth to this. Several active site residues have to move "outward" by 0.5 Angstroms or so to accommodate the inhibitor. Other residues move "inward." There are, however, other factors that come into play. In particular, the side chain of His310 rotates by 134 degrees into a different position where (together with Glu128 and Tyr195) it coordinates a metal ion not seen at this site in the native enzyme.
Figures
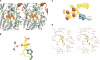


References
-
- Addlagatta A., Hu X., Liu J.O., Matthews B.W. 2005. Structural basis for the functional differences between Type I and Type II human methionine aminopeptidases. Biochemistry 44: 14741–14749. - PubMed
-
- Bergers G., Javaherian K., Lo K.M., Folkman J., Hanahan D. 1999. Effects of angiogenesis inhibitors on multistage carcinogenesis in mice. Science 284: 808–812. - PubMed
-
- Bhargava P., Marshall J.L., Rizvi N., Dahut W., Yoe J., Figuera M., Phipps K., Ong V.S., Kato A., Hawkins M.J. 1999. A Phase I and pharmacokinetic study of TNP-470 administered weekly to patients with advanced cancer. Clin. Cancer Res. 5: 1989–1995. - PubMed
-
- Brünger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S. et al. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54: 905–921. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

