Mouse chromosome engineering for modeling human disease
- PMID: 16824018
- PMCID: PMC2597817
- DOI: 10.1146/annurev.genom.7.080505.115741
Mouse chromosome engineering for modeling human disease
Abstract
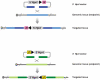
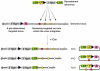
Chromosomal rearrangements are frequently in humans and can be disease-associated or phenotypically neutral. Recent technological advances have led to the discovery of copy-number changes previously undetected by cytogenetic techniques. To understand the genetic consequences of such genomic changes, these mutations need to be modeled in experimentally tractable systems. The mouse is an excellent organism for this analysis because of its biological and genetic similarity to humans, and the ease with which its genome can be manipulated. Through chromosome engineering, defined rearrangements can be introduced into the mouse genome. The resulting mouse models are leading to a better understanding of the molecular and cellular basis of dosage alterations in human disease phenotypes, in turn opening new diagnostic and therapeutic opportunities.
Figures




Similar articles
-
Chromosome engineering in ES cells.Methods Mol Biol. 2009;530:49-77. doi: 10.1007/978-1-59745-471-1_4. Methods Mol Biol. 2009. PMID: 19266329 Review.
-
Mice and humans: chromosome engineering and its application to functional genomics.Acta Biochim Pol. 2002;49(3):553-69. Acta Biochim Pol. 2002. PMID: 12422227 Review.
-
Two patterns of genome organization in mammals: the chromosomal distribution of duplicate genes in human and mouse.Mol Biol Evol. 2004 Jun;21(6):1008-13. doi: 10.1093/molbev/msh076. Epub 2004 Feb 12. Mol Biol Evol. 2004. PMID: 14963098
-
Engineering chromosomal rearrangements in mice.Nat Rev Genet. 2001 Oct;2(10):780-90. doi: 10.1038/35093564. Nat Rev Genet. 2001. PMID: 11584294 Review.
-
Genome rearrangements in mammalian evolution: lessons from human and mouse genomes.Genome Res. 2003 Jan;13(1):37-45. doi: 10.1101/gr.757503. Genome Res. 2003. PMID: 12529304 Free PMC article.
Cited by
-
Transgenic mouse technology: principles and methods.Methods Mol Biol. 2009;590:335-62. doi: 10.1007/978-1-60327-378-7_22. Methods Mol Biol. 2009. PMID: 19763515 Free PMC article.
-
Using genetic mouse models to gain insight into glaucoma: Past results and future possibilities.Exp Eye Res. 2015 Dec;141:42-56. doi: 10.1016/j.exer.2015.06.019. Epub 2015 Jun 24. Exp Eye Res. 2015. PMID: 26116903 Free PMC article. Review.
-
A mouse model of lamellar intrastromal femtosecond laser keratotomy: ultra-structural, inflammatory, and wound healing responses.Mol Vis. 2011;17:3005-12. Epub 2011 Nov 17. Mol Vis. 2011. PMID: 22171154 Free PMC article.
-
Following the genes: a framework for animal modeling of psychiatric disorders.BMC Biol. 2011 Nov 11;9:76. doi: 10.1186/1741-7007-9-76. BMC Biol. 2011. PMID: 22078115 Free PMC article. Review.
-
Mouse models of genomic syndromes as tools for understanding the basis of complex traits: an example with the smith-magenis and the potocki-lupski syndromes.Curr Genomics. 2009 Jun;10(4):259-68. doi: 10.2174/138920209788488508. Curr Genomics. 2009. PMID: 19949547 Free PMC article.
References
LITERATURE CITED
-
- Adams DJ, Biggs PJ, Cox T, Davies R, van der Weyden L, et al. Mutagenic insertion and chromosome engineering resource (MICER) Nat. Genet. 2004;36:867–71. - PubMed
-
- Adams DJ, Dermitzakis ET, Cox T, Smith J, Davies R, et al. Complex haplotypes, copy number polymorphisms and coding variation in two recently divergent mouse strains. Nat. Genet. 2005;37:532–36. - PubMed
-
- Albertson DG, Pinkel D. Genomic microarrays in human genetic disease and cancer. Hum. Mol. Genet. 2003;12(Spec. No. 2):R145–52. - PubMed
-
- Baldini A. Dissecting contiguous gene defects: TBX1. Curr. Opin. Genet. Dev. 2005;15:279–84. - PubMed
RELATED RESOURCE
-
- Sharp AJ, Cheng Z, Eichler E. Structural variation of the human genome. Annu. Rev. Genomics Hum. Genet. 2006;7:407–42. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

