A quantitative reverse transcriptase-polymerase chain reaction assay to identify metastatic carcinoma tissue of origin
- PMID: 16825504
- PMCID: PMC1867609
- DOI: 10.2353/jmoldx.2006.050136
A quantitative reverse transcriptase-polymerase chain reaction assay to identify metastatic carcinoma tissue of origin
Abstract
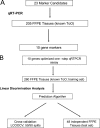
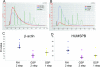
Identifying the primary site in patients with metastatic carcinoma of unknown primary origin can enable more specific therapeutic regimens and may prolong survival. Twenty-three putative tissue-specific markers for lung, colon, pancreatic, breast, prostate, and ovarian carcinomas were nominated by querying a gene expression profile database and by performing a literature search. Ten of these marker candidates were then selected based on validation by reverse transcriptase-polymerase chain reaction (RT-PCR) on 205 formalin-fixed, paraffin-embedded metastatic carcinoma specimens originating from these six and from other cancer types. Next, we optimized the RNA isolation and quantitative RT-PCR methods for these 10 markers and applied the quantitative RT-PCR assay to a set of 260 metastatic tumors. We then built a gene-based algorithm that predicted the tissue of origin of metastatic carcinomas with an overall leave-one-out cross-validation accuracy of 78%. Lastly, our assay demonstrated an accuracy of 76% when tested on an independent set of 48 metastatic samples, 37 of which were either a known primary or initially presented as carcinoma of unknown primary but were subsequently resolved.
Figures


References
-
- Ghosh L, Dahut W, Kakar S, Posadas EM, Torres CG, Cancel-Santiago R, Ghosh BC. Management of patients with metastatic cancer of unknown primary. Curr Probl Surg. 2005;42:12–66. - PubMed
-
- Mintzer DM, Warhol M, Martin A-M, Greene G. Cancer of unknown primary: changing approaches, a multidisciplinary case presentation from the Joan Karnell Cancer Center of Pennsylvania Hospital. Oncologist. 2004;9:330–338. - PubMed
-
- Greco FA, Rodriguez GI, Shaffer DW, Hermann R, Litchy S, Yardley DA, Burris HA, III, Morrissey LH, Erland JB, Hainsworth JD. Carcinoma of unknown primary site: sequential treatment with paclitaxel/carboplatin/etoposide and gemcitabine/irinotecan. A Minnie Pearl cancer research network phase II trial. Oncologist. 2004;9:644–652. - PubMed
-
- Brown RW, Campagna LB, Dunn JK, Cagle PT. Immunohistochemical identification of tumor markers in metastatic adenocarcinoma: a diagnostic adjunct in the determination of primary site. Am J Clin Pathol. 1997;107:12–19. - PubMed
-
- DeYoung BR, Wick MR. Immunohistologic evaluation of metastatic carcinomas of unknown origin: an algorithmic approach. Semin Diagn Pathol. 2000;17:184–193. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

