Mutation in TRMU related to transfer RNA modification modulates the phenotypic expression of the deafness-associated mitochondrial 12S ribosomal RNA mutations
- PMID: 16826519
- PMCID: PMC1559489
- DOI: 10.1086/506389
Mutation in TRMU related to transfer RNA modification modulates the phenotypic expression of the deafness-associated mitochondrial 12S ribosomal RNA mutations
Abstract
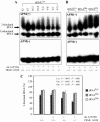
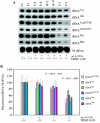
The human mitochondrial 12S ribosomal RNA (rRNA) A1555G mutation has been associated with aminoglycoside-induced and nonsyndromic deafness in many families worldwide. Our previous investigation revealed that the A1555G mutation is a primary factor underlying the development of deafness but is not sufficient to produce a deafness phenotype. However, it has been proposed that nuclear-modifier genes modulate the phenotypic manifestation of the A1555G mutation. Here, we identified the nuclear-modifier gene TRMU, which encodes a highly conserved mitochondrial protein related to transfer RNA (tRNA) modification. Genotyping analysis of TRMU in 613 subjects from 1 Arab-Israeli kindred, 210 European (Italian pedigrees and Spanish pedigrees) families, and 31 Chinese pedigrees carrying the A1555G or the C1494T mutation revealed a missense mutation (G28T) altering an invariant amino acid residue (A10S) in the evolutionarily conserved N-terminal region of the TRMU protein. Interestingly, all 18 Arab-Israeli/Italian-Spanish matrilineal relatives carrying both the TRMU A10S and 12S rRNA A1555G mutations exhibited prelingual profound deafness. Functional analysis showed that this mutation did not affect importation of TRMU precursors into mitochondria. However, the homozygous A10S mutation leads to a marked failure in mitochondrial tRNA metabolisms, specifically reducing the steady-state levels of mitochondrial tRNA. As a consequence, these defects contribute to the impairment of mitochondrial-protein synthesis. Resultant biochemical defects aggravate the mitochondrial dysfunction associated with the A1555G mutation, exceeding the threshold for expressing the deafness phenotype. These findings indicate that the mutated TRMU, acting as a modifier factor, modulates the phenotypic manifestation of the deafness-associated 12S rRNA mutations.
Figures
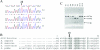



References
Web Resources
-
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for the complete TRMU sequences [accession number AF448221])
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for deafness-associated mutations in 12S rRNA)
References
-
- Estivill X, Govea N, Barceló A, Perelló E, Badenas C, Romero E, Moral L, Scozzari R, D’Urbano L, Zeviani M, Torroni A (1998) Familial progressive sensorineural deafness is mainly due to the mtDNA A1555G mutation and is enhanced by treatment with aminoglycosides. Am J Hum Genet 62:27–35 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

