Methylation of neutral endopeptidase 24.11 promoter in rat hepatocellular carcinoma
- PMID: 16827801
- PMCID: PMC11158973
- DOI: 10.1111/j.1349-7006.2006.00227.x
Methylation of neutral endopeptidase 24.11 promoter in rat hepatocellular carcinoma
Abstract
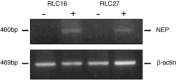
Neutral endopeptidase 24.11 (NEP), a cell-surface enzyme expressed by epithelial cells that cleaves and inactivates biologically active small peptides, is downregulated in various cancers. NEP is encoded by a gene that contains a CpG island in the promoter region, whose hypermethylation appears related to decreased expression. Altered expression of NEP has also been reported in human hepatocellular carcinoma (HCC), suggesting its possible role in hepatocarcinogenesis. To elucidate the status of NEP in HCC, methylation in the promoter region of the gene that encodes NEP in male Fischer 344 rats with HCC, induced by a choline-deficient, l-amino acid-defined diet, was investigated by methylation-specific polymerase chain reaction, combined bisulfite restriction analysis, and bisulfite genomic sequencing. These analyses together showed the promoter to be frequently methylated in HCC in contrast to its unmethylated status in normal liver, the degree of methylation being inversely related to the level of mRNA expression evaluated by reverse transcription-polymerase chain reaction (P = 0.031). In two rat liver cell lines, RLC-16 and RLC-27, the promoter was heavily methylated and NEP mRNA expression was negative. However, administration of 5-aza-2'-deoxycytidine caused NEP expression, suggesting that methylation of CpG is a factor regulating transcriptional expression. Together with the data from microarray analyses performed previously using the same animal model, the current results suggest that reduced expression of NEP or other ectopeptidases could impact on molecules involved in signal-transducing systems, including G-protein coupled receptors, via modified turnover of extracellularly active small peptides.
Figures



Similar articles
-
Methylation of the neutral endopeptidase gene promoter in human prostate cancers.Clin Cancer Res. 2000 May;6(5):1664-70. Clin Cancer Res. 2000. PMID: 10815884
-
Demethylation by 5-aza-2'-deoxycytidine in colorectal cancer cells targets genomic DNA whilst promoter CpG island methylation persists.BMC Cancer. 2010 Jul 12;10:366. doi: 10.1186/1471-2407-10-366. BMC Cancer. 2010. PMID: 20618997 Free PMC article.
-
XAF1 is frequently methylated in human esophageal cancer.World J Gastroenterol. 2012 Jun 14;18(22):2844-9. doi: 10.3748/wjg.v18.i22.2844. World J Gastroenterol. 2012. PMID: 22719195 Free PMC article.
-
Aberrant methylation and downregulation of sall3 in human hepatocellular carcinoma.World J Gastroenterol. 2012 Jun 7;18(21):2719-26. doi: 10.3748/wjg.v18.i21.2719. World J Gastroenterol. 2012. PMID: 22690083 Free PMC article.
-
The relationship of MITF gene expression and promoter methylation with plumage colour in quail.Br Poult Sci. 2024 Jun;65(3):259-264. doi: 10.1080/00071668.2024.2326962. Epub 2024 Apr 5. Br Poult Sci. 2024. PMID: 38578288 Review.
Cited by
-
Neprilysin gene expression requires binding of the amyloid precursor protein intracellular domain to its promoter: implications for Alzheimer disease.EMBO Rep. 2009 Jan;10(1):94-100. doi: 10.1038/embor.2008.222. Epub 2008 Dec 5. EMBO Rep. 2009. PMID: 19057576 Free PMC article.
-
Disturbance of DNA methylation patterns in the early phase of hepatocarcinogenesis induced by a choline-deficient L-amino acid-defined diet in rats.Cancer Sci. 2007 Sep;98(9):1318-22. doi: 10.1111/j.1349-7006.2007.00564.x. Epub 2007 Jul 19. Cancer Sci. 2007. PMID: 17640295 Free PMC article.
-
Involvement of mutation-based inhibition of beta-catenin phosphorylation at Ser33 in the malignant progression of lung (pre)neoplastic lesions induced by N-nitrosobis(2-hydroxypropyl)amine in male Fischer 344 rats.Lung. 2007 Sep-Oct;185(5):271-278. doi: 10.1007/s00408-007-9017-y. Epub 2007 Jul 18. Lung. 2007. PMID: 17639448
-
Polyhydroxycurcuminoids but not curcumin upregulate neprilysin and can be applied to the prevention of Alzheimer's disease.Sci Rep. 2016 Jul 13;6:29760. doi: 10.1038/srep29760. Sci Rep. 2016. PMID: 27407064 Free PMC article.
-
Prognostic significance of the combined expression of neutral endopeptidase and dipeptidyl peptidase IV in intrahepatic cholangiocarcinoma patients after surgery resection.Onco Targets Ther. 2014 Feb 17;7:297-304. doi: 10.2147/OTT.S57355. eCollection 2014. Onco Targets Ther. 2014. PMID: 24570591 Free PMC article.
References
-
- Nanus DM. Of peptides and peptidases: the role of cell surface peptidases in cancer. Clin Cancer Res 2003; 9: 6307–9. - PubMed
-
- Cohen AJ, Franklin WA, Magill C, Sorenson J, Miller YE. Low neutral endopeptidase levels in bronchoalveolar lavage fluid of lung cancer patients. Am J Respir Crit Care Med 1999; 159: 907–10. - PubMed
-
- Papandreou CN, Usmani B, Geng Y et al. Neutral endopeptidase 24.11 loss in metastatic human prostate cancer contributes to androgen‐independent progression. Nat Med 1998; 4: 50–7. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

