Crystal structure of staphylococcal enterotoxin I (SEI) in complex with a human major histocompatibility complex class II molecule
- PMID: 16829512
- PMCID: PMC2730046
- DOI: 10.1074/jbc.M603969200
Crystal structure of staphylococcal enterotoxin I (SEI) in complex with a human major histocompatibility complex class II molecule
Abstract
Superantigens are bacterial or viral proteins that elicit massive T cell activation through simultaneous binding to major histocompatibility complex (MHC) class II and T cell receptors. This activation results in uncontrolled release of inflammatory cytokines, causing toxic shock. A remarkable property of superantigens, which distinguishes them from T cell receptors, is their ability to interact with multiple MHC class II alleles independently of MHC-bound peptide. Previous crystallographic studies have shown that staphylococcal and streptococcal superantigens belonging to the zinc family bind to a high affinity site on the class II beta-chain. However, the basis for promiscuous MHC recognition by zinc-dependent superantigens is not obvious, because the beta-chain is polymorphic and the MHC-bound peptide forms part of the binding interface. To understand how zinc-dependent superantigens recognize MHC, we determined the crystal structure, at 2.0 A resolution, of staphylococcal enterotoxin I bound to the human class II molecule HLA-DR1 bearing a peptide from influenza hemagglutinin. Interactions between the superantigen and DR1 beta-chain are mediated by a zinc ion, and 22% of the buried surface of peptide.MHC is contributed by the peptide. Comparison of the staphylococcal enterotoxin I.peptide.DR1 structure with ones determined previously revealed that zinc-dependent superantigens achieve promiscuous binding to MHC by targeting conservatively substituted residues of the polymorphic beta-chain. Additionally, these superantigens circumvent peptide specificity by engaging MHC-bound peptides at their conformationally conserved N-terminal regions while minimizing sequence-specific interactions with peptide residues to enhance cross-reactivity.
Figures

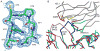

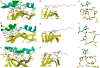
Similar articles
-
Crystal structure of a superantigen bound to MHC class II displays zinc and peptide dependence.EMBO J. 2001 Jul 2;20(13):3306-12. doi: 10.1093/emboj/20.13.3306. EMBO J. 2001. PMID: 11432818 Free PMC article.
-
A recombinant single-chain human class II MHC molecule (HLA-DR1) as a covalently linked heterotrimer of alpha chain, beta chain, and antigenic peptide, with immunogenicity in vitro and reduced affinity for bacterial superantigens.Eur J Immunol. 1997 Aug;27(8):1933-41. doi: 10.1002/eji.1830270817. Eur J Immunol. 1997. PMID: 9295029
-
Staphylococcal enterotoxin A has two cooperative binding sites on major histocompatibility complex class II.J Exp Med. 1995 Sep 1;182(3):711-20. doi: 10.1084/jem.182.3.711. J Exp Med. 1995. PMID: 7650479 Free PMC article.
-
Structure-function studies of T-cell receptor-superantigen interactions.Immunol Rev. 1998 Jun;163:177-86. doi: 10.1111/j.1600-065x.1998.tb01196.x. Immunol Rev. 1998. PMID: 9700510 Review.
-
Superantigens. Gazing into the crystal ball.Curr Biol. 1995 Mar 1;5(3):235-7. doi: 10.1016/s0960-9822(95)00047-9. Curr Biol. 1995. PMID: 7780728 Review.
Cited by
-
Docking-Based Prediction of Peptide Binding to MHC Proteins.Methods Mol Biol. 2023;2673:237-249. doi: 10.1007/978-1-0716-3239-0_17. Methods Mol Biol. 2023. PMID: 37258919
-
A novel loop domain in superantigens extends their T cell receptor recognition site.J Mol Biol. 2007 Aug 3;371(1):210-21. doi: 10.1016/j.jmb.2007.05.038. Epub 2007 May 18. J Mol Biol. 2007. PMID: 17560605 Free PMC article.
-
Superantigens, a Paradox of the Immune Response.Toxins (Basel). 2022 Nov 18;14(11):800. doi: 10.3390/toxins14110800. Toxins (Basel). 2022. PMID: 36422975 Free PMC article. Review.
-
Staphylococcal superantigens in colonization and disease.Front Cell Infect Microbiol. 2012 Apr 17;2:52. doi: 10.3389/fcimb.2012.00052. eCollection 2012. Front Cell Infect Microbiol. 2012. PMID: 22919643 Free PMC article. Review.
-
The systemic and pulmonary immune response to staphylococcal enterotoxins.Toxins (Basel). 2010 Jul;2(7):1898-912. doi: 10.3390/toxins2071898. Epub 2010 Jul 21. Toxins (Basel). 2010. PMID: 22069664 Free PMC article. Review.
References
-
- Scherer MT, Ignatowicz L, Winslow GM, Kappler JW, Marrack P. Annu Rev Cell Biol. 1993;9:101–128. - PubMed
-
- Li H, Llera A, Malchiodi EL, Mariuzza RA. Annu Rev Immunol. 1999;17:435–466. - PubMed
-
- McCormick JK, Yarwood JM, Schlievert PM. Annu Rev Microbiol. 2001;55:77–104. - PubMed
-
- Sundberg EJ, Li Y, Mariuzza RA. Curr Opin Immunol. 2002;14:36–44. - PubMed
-
- Bohach GA, Fast DJ, Nelson RD, Schlievert PM. Crit Rev Microbiol. 1990;17:251–272. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

