The C-terminal zinc finger of UvrA does not bind DNA directly but regulates damage-specific DNA binding
- PMID: 16829526
- PMCID: PMC2396232
- DOI: 10.1074/jbc.M603093200
The C-terminal zinc finger of UvrA does not bind DNA directly but regulates damage-specific DNA binding
Abstract
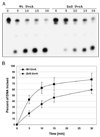
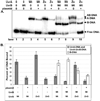
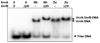
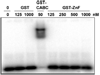
In prokaryotic nucleotide excision repair, UvrA recognizes DNA perturbations and recruits UvrB for the recognition and processing steps in the reaction. One of the most remarkable aspects of UvrA is that it can recognize a wide range of DNA lesions that differ in chemistry and structure. However, how UvrA interacts with DNA is unknown. To examine the role that the UvrA C-terminal zinc finger domain plays in DNA binding, an eleven amino acid deletion was constructed (ZnG UvrA). Biochemical characterization of the ZnG UvrA protein was carried out using UvrABC DNA incision, DNA binding and ATPase assays. Although ZnG UvrA was able to bind dsDNA slightly better than wild-type UvrA, the ZnG UvrA mutant only supported 50-75% of wild type incision. Surprisingly, the ZnG UvrA mutant, while retaining its ability to bind dsDNA, did not support damage-specific binding. Furthermore, this mutant protein only provided 10% of wild-type Bca UvrA complementation for UV survival of an uvrA deletion strain. In addition, ZnG UvrA failed to stimulate the UvrB DNA damage-associated ATPase activity. Electrophoretic mobility shift analysis was used to monitor UvrB loading onto damaged DNA with wild-type UvrA or ZnG UvrA. The ZnG UvrA protein showed a 30-60% reduction in UvrB loading as compared with the amount of UvrB loaded by wild-type UvrA. These data demonstrate that the C-terminal zinc finger of UvrA is required for regulation of damage-specific DNA binding.
Figures






References
-
- Van Houten B, Croteau DL, DellaVecchia MJ, Wang H, Kisker C. Mutat. Res. 2005;577:92–117. - PubMed
-
- Truglio JJ, Croteau DL, Van Houten B, Kisker C. Chem. Rev. 2006;106:233–252. - PubMed
-
- Croteau DL, Dellavecchia MJ, Skorvaga M, Van Houten B. In: DNA Damage Recognition. Wolfram S, Kow YW, Doetsch PW, editors. Boca Raton, FL: Taylor & Francis; 2005. pp. 111–138.
-
- Reardon JT, Sancar A. Cell Cycle. 2004;3:141–144. - PubMed
-
- Doolittle RF, Johnson MS, Husain I, Van Houten B, Thomas DC, Sancar A. Nature. 1986;323:451–453. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

