Size-dependent diffusion of membrane inclusions
- PMID: 16829562
- PMCID: PMC1562383
- DOI: 10.1529/biophysj.106.087031
Size-dependent diffusion of membrane inclusions
Abstract
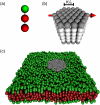
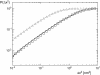
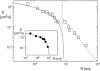
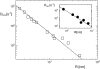
Experimentally determined diffusion constants are often used to elucidate the size and oligomeric state of membrane proteins and domains. This approach critically relies on the knowledge of the size-dependence of diffusion. We have used mesoscopic simulations to thoroughly quantify the size-dependent diffusion properties of membrane inclusions. For small radii R, we find that the lateral diffusion coefficient D is well described by the Saffman-Delbrück relation, which predicts a logarithmic decrease of D with R. However, beyond a critical radius Rc approximately hetam/(2etac) (h, bilayer thickness; etam/c, viscosity of the membrane/surrounding solvent) we observe significant deviations and the emergence of an asymptotic scaling D approximately 1/R2. The latter originates from the asymptotic hydrodynamics and the inclusion's internal degrees of freedom that become particularly relevant on short timescales. In contrast to the lateral diffusion, the size dependence of the rotational diffusion constant Dr follows the predicted hydrodynamic scaling Dr approximately 1/R2 over the entire range of sizes studied here.
Figures
Similar articles
-
Influence of hydrophobic mismatching on membrane protein diffusion.Biophys J. 2008 Aug;95(3):L25-7. doi: 10.1529/biophysj.108.136069. Epub 2008 May 23. Biophys J. 2008. PMID: 18502792 Free PMC article.
-
Translational diffusion in lipid membranes beyond the Saffman-Delbruck approximation.Biophys J. 2008 Mar 1;94(5):L41-3. doi: 10.1529/biophysj.107.126565. Epub 2008 Jan 11. Biophys J. 2008. PMID: 18192354 Free PMC article.
-
Mobility in geometrically confined membranes.Proc Natl Acad Sci U S A. 2011 Aug 2;108(31):12605-10. doi: 10.1073/pnas.1102646108. Epub 2011 Jul 18. Proc Natl Acad Sci U S A. 2011. PMID: 21768336 Free PMC article.
-
Resolving the kinetics of lipid, protein and peptide diffusion in membranes.Mol Membr Biol. 2012 Aug;29(5):118-43. doi: 10.3109/09687688.2012.678018. Epub 2012 May 14. Mol Membr Biol. 2012. PMID: 22582994 Review.
-
Non-Brownian diffusion in lipid membranes: Experiments and simulations.Biochim Biophys Acta. 2016 Oct;1858(10):2451-2467. doi: 10.1016/j.bbamem.2016.01.022. Epub 2016 Jan 28. Biochim Biophys Acta. 2016. PMID: 26826272 Review.
Cited by
-
Membrane fluidity and lipid order in ternary giant unilamellar vesicles using a new bodipy-cholesterol derivative.Biophys J. 2009 Apr 8;96(7):2696-708. doi: 10.1016/j.bpj.2008.12.3922. Biophys J. 2009. PMID: 19348752 Free PMC article.
-
Rapid Enrichment of a Native Multipass Transmembrane Protein via Cell Membrane Electrophoresis through Buffer pH and Ionic Strength Adjustment.J Am Chem Soc. 2024 May 1;146(17):11634-11647. doi: 10.1021/jacs.3c13579. Epub 2024 Apr 17. J Am Chem Soc. 2024. PMID: 38628144 Free PMC article.
-
Phase separation in biological membranes: integration of theory and experiment.Annu Rev Biophys. 2010;39:207-26. doi: 10.1146/annurev.biophys.093008.131238. Annu Rev Biophys. 2010. PMID: 20192775 Free PMC article. Review.
-
Assembly formation of minor dihydrosphingomyelin in sphingomyelin-rich ordered membrane domains.Sci Rep. 2020 Jul 16;10(1):11794. doi: 10.1038/s41598-020-68688-7. Sci Rep. 2020. PMID: 32678223 Free PMC article.
-
Quantitative microscopy reveals dynamics and fate of clustered IRE1α.Proc Natl Acad Sci U S A. 2020 Jan 21;117(3):1533-1542. doi: 10.1073/pnas.1915311117. Epub 2019 Dec 23. Proc Natl Acad Sci U S A. 2020. PMID: 31871156 Free PMC article.
References
-
- Singer, S. J., and G. L. Nicholson. 1972. The fluid mosaic model of the structure of cell membranes. Science. 175:720–731. - PubMed
-
- Engelman, D. M. 2005. Membranes are more mosaic than fluid. Nature. 438:578–580. - PubMed
-
- Simons, K., and E. Ikonen. 1997. Functional rafts in cell membranes. Nature. 119:569–572. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

