A molecular correlate to the Gleason grading system for prostate adenocarcinoma
- PMID: 16829574
- PMCID: PMC1544162
- DOI: 10.1073/pnas.0603678103
A molecular correlate to the Gleason grading system for prostate adenocarcinoma
Abstract
Adenocarcinomas of the prostate can be categorized into tumor grades based on the extent to which the cancers histologically resemble normal prostate glands. Because grades are surrogates of intrinsic tumor behavior, characterizing the molecular phenotype of grade is of potential clinical importance. To identify molecular alterations underlying prostate cancer grades, we used microdissection to obtain specific cohorts of cancer cells corresponding to the most common Gleason patterns (patterns 3, 4, and 5) from 29 radical prostatectomy samples. We paired each cancer sample with matched benign lumenal prostate epithelial cells and profiled transcript abundance levels by microarray analysis. We identified an 86-gene model capable of distinguishing low-grade (pattern 3) from high-grade (patterns 4 and 5) cancers. This model performed with 76% accuracy when applied to an independent set of 30 primary prostate carcinomas. Using tissue microarrays comprising >800 prostate samples, we confirmed a significant association between high levels of monoamine oxidase A expression and poorly differentiated cancers by immunohistochemistry. We also confirmed grade-associated levels of defender against death (DAD1) protein and HSD17 beta4 transcripts by immunohistochemistry and quantitative RT-PCR, respectively. The altered expression of these genes provides functional insights into grade-associated features of therapy resistance and tissue invasion. Furthermore, in identifying a profile of 86 genes that distinguish high- from low-grade carcinomas, we have generated a set of potential targets for modulating the development and progression of the lethal prostate cancer phenotype.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures
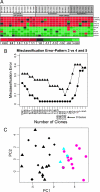
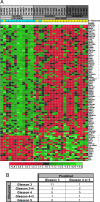

Comment in
-
Re: A molecular correlate to the Gleason grading system for prostate adenocarcinoma.Eur Urol. 2007 Mar;51(3):851-2. doi: 10.1016/j.eururo.2006.12.006. Eur Urol. 2007. PMID: 17421063 No abstract available.
Similar articles
-
Usual and unusual histologic patterns of high Gleason score 8 to 10 adenocarcinoma of the prostate in needle biopsy tissue.Am J Surg Pathol. 2012 Jun;36(6):900-7. doi: 10.1097/PAS.0b013e3182495dee. Am J Surg Pathol. 2012. PMID: 22367295
-
Gene-expression analysis of gleason grade 3 tumor glands embedded in low- and high-risk prostate cancer.Oncotarget. 2016 Jun 21;7(25):37846-37856. doi: 10.18632/oncotarget.9344. Oncotarget. 2016. PMID: 27191985 Free PMC article.
-
Minimum altered regions in early prostate cancer progression identified by high resolution whole genome tiling path BAC array comparative hybridization.Prostate. 2009 Jun 15;69(9):961-75. doi: 10.1002/pros.20949. Prostate. 2009. PMID: 19267368
-
Pathological and molecular aspects of prostate cancer.Lancet. 2003 Mar 15;361(9361):955-64. doi: 10.1016/S0140-6736(03)12779-1. Lancet. 2003. PMID: 12648986 Review.
-
Histopathology of Prostate Cancer.Cold Spring Harb Perspect Med. 2017 Oct 3;7(10):a030411. doi: 10.1101/cshperspect.a030411. Cold Spring Harb Perspect Med. 2017. PMID: 28389514 Free PMC article. Review.
Cited by
-
Regulation of 17β-hydroxysteroid dehydrogenases in cancer: regulating steroid receptor at pre-receptor stage.J Physiol Biochem. 2012 Sep;68(3):461-73. doi: 10.1007/s13105-012-0155-1. Epub 2012 Feb 29. J Physiol Biochem. 2012. PMID: 22374586 Review.
-
A regulatory feedback loop between Ca2+/calmodulin-dependent protein kinase kinase 2 (CaMKK2) and the androgen receptor in prostate cancer progression.J Biol Chem. 2012 Jul 13;287(29):24832-43. doi: 10.1074/jbc.M112.370783. Epub 2012 May 31. J Biol Chem. 2012. PMID: 22654108 Free PMC article.
-
PPP2R2C loss promotes castration-resistance and is associated with increased prostate cancer-specific mortality.Mol Cancer Res. 2013 Jun;11(6):568-78. doi: 10.1158/1541-7786.MCR-12-0710. Epub 2013 Mar 14. Mol Cancer Res. 2013. PMID: 23493267 Free PMC article.
-
Factor interaction analysis for chromosome 8 and DNA methylation alterations highlights innate immune response suppression and cytoskeletal changes in prostate cancer.Mol Cancer. 2007 Feb 5;6:14. doi: 10.1186/1476-4598-6-14. Mol Cancer. 2007. PMID: 17280610 Free PMC article.
-
Patterns of indolence in prostate cancer (Review).Exp Ther Med. 2022 May;23(5):351. doi: 10.3892/etm.2022.11278. Epub 2022 Mar 28. Exp Ther Med. 2022. PMID: 35493432 Free PMC article. Review.
References
-
- Partin A. W., Kattan M. W., Subong E. N., Walsh P. C., Wojno K. J., Oesterling J. E., Scardino P. T., Pearson J. D. J. Am. Med. Assoc. 1997;277:1445–1451. - PubMed
-
- Gleason D. F., Mellinger G. T. J. Urol. 1974;111:58–64. - PubMed
-
- Epstein J. I., Allsbrook W. C., Jr., Amin M. B., Egevad L. L. Am. J. Surg. Pathol. 2005;29:1228–1242. - PubMed
-
- Epstein J. I. Am. J. Surg. Pathol. 2000;24:477–478. - PubMed
-
- Albertsen P. C., Hanley J. A., Barrows G. H., Penson D. F., Kowalczyk P. D., Sanders M. M., Fine J. J. Natl. Cancer Inst. 2005;97:1248–1253. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

