Rapid genetic grouping of factor h-binding protein (genome-derived neisserial antigen 1870), a promising group B meningococcal vaccine candidate
- PMID: 16829612
- PMCID: PMC1489572
- DOI: 10.1128/CVI.00097-06
Rapid genetic grouping of factor h-binding protein (genome-derived neisserial antigen 1870), a promising group B meningococcal vaccine candidate
Abstract
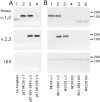
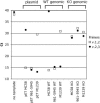
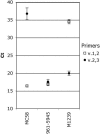
The most important antigen component of a promising multicomponent group B meningococcal recombinant protein vaccine is based on genome-derived neisserial antigen 1870, which recently was renamed factor H-binding protein (FHBP) to reflect one of its critical functions as a complement regulatory protein. Neisseria meningitidis strains can be subdivided into three FHBP variant groups based on divergence of FHBP amino acid sequences. Within each variant group, amino acid sequences are >90% conserved. To develop an FHBP-based group B vaccine, it is important to know the distribution of FHBP variant 1, 2, and 3 strains in different geographic regions, since antibodies against FHBP are bactericidal against strains within the homologous group but show minimal activity against strains from other groups. We have devised a high-throughput, quantitative PCR-based method that allows rapid and precise assignment of FHBP genes into each of the three major variant lineages. Among 48 group B isolates from patients hospitalized in California in 2003 to 2004, 83%, 13%, and 4%, respectively, had variant 1, 2, and 3 genes. Thus, a vaccine based on the variant 1 protein has the potential to prevent the majority of cases of group B disease. The quantitative PCR-based method will be useful for determining and monitoring the prevalence of meningococcal isolates with genes encoding different FHBP variant proteins. The technique also is suitable for monitoring variation of genes encoding other protein antigens targeted for vaccination.
Figures

References
-
- Brody, J. R., and S. E. Kern. 2004. Sodium boric acid: a Tris-free, cooler conductive medium for DNA electrophoresis. BioTechniques 36:214-216. - PubMed
-
- Cartwright, K., N. Noah, and H. Peltola. 2001. Meningococcal disease in Europe: epidemiology, mortality, and prevention with conjugate vaccines. Report of a European advisory board meeting Vienna, Austria, 6-8 October, 2000. Vaccine 19:4347-4356. - PubMed
-
- Comanducci, M., S. Bambini, B. Brunelli, J. Adu-Bobie, B. Arico, B. Capecchi, M. M. Giuliani, V. Masignani, L. Santini, S. Savino, D. M. Granoff, D. A. Caugant, M. Pizza, R. Rappuoli, and M. Mora. 2002. NadA, a novel vaccine candidate of Neisseria meningitidis. J. Exp. Med. 195:1445-1454. - PMC - PubMed
-
- Crowe, B. A., R. A. Wall, B. Kusecek, B. Neumann, T. Olyhoek, H. Abdillahi, M. Hassan-King, B. M. Greenwood, J. T. Poolman, and M. Achtman. 1989. Clonal and variable properties of Neisseria meningitidis isolated from cases and carriers during and after an epidemic in The Gambia, West Africa. J. Infect. Dis. 159:686-700. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

