Yellow flowers generated by expression of the aurone biosynthetic pathway
- PMID: 16832053
- PMCID: PMC1544175
- DOI: 10.1073/pnas.0604246103
Yellow flowers generated by expression of the aurone biosynthetic pathway
Abstract
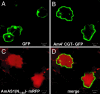
Flower color is most often conferred by colored flavonoid pigments. Aurone flavonoids confer a bright yellow color on flowers such as snapdragon (Antirrhinum majus) and dahlia (Dahlia variabilis). A. majus aureusidin synthase (AmAS1) was identified as the key enzyme that catalyzes aurone biosynthesis from chalcones, but transgenic flowers overexpressing AmAS1 gene failed to produce aurones. Here, we report that chalcone 4'-O-glucosyltransferase (4'CGT) is essential for aurone biosynthesis and yellow coloration in vivo. Coexpression of the Am4'CGT and AmAS1 genes was sufficient for the accumulation of aureusidin 6-O-glucoside in transgenic flowers (Torenia hybrida). Furthermore, their coexpression combined with down-regulation of anthocyanin biosynthesis by RNA interference (RNAi) resulted in yellow flowers. An Am4'CGT-GFP chimeric protein localized in the cytoplasm, whereas the AmAS1(N1-60)-RFP chimeric protein was localized to the vacuole. We therefore conclude that chalcones are 4'-O-glucosylated in the cytoplasm, their 4'-O-glucosides transported to the vacuole, and therein enzymatically converted to aurone 6-O-glucosides. This metabolic pathway is unique among the known examples of flavonoid, including anthocyanin biosynthesis because, for all other compounds, the carbon backbone is completed before transport to the vacuole. Our findings herein not only demonstrate the biochemical basis of aurone biosynthesis but also open the way to engineering yellow flowers for major ornamental species lacking this color variant.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures



Similar articles
-
Generation of Yellow Flowers of the Japanese Morning Glory by Engineering Its Flavonoid Biosynthetic Pathway toward Aurones.Plant Cell Physiol. 2019 Aug 1;60(8):1871-1879. doi: 10.1093/pcp/pcz101. Plant Cell Physiol. 2019. PMID: 31135027
-
Altered leaf colour is associated with increased superoxide-scavenging activity in aureusidin-producing transgenic plants.Plant Biotechnol J. 2012 Dec;10(9):1046-55. doi: 10.1111/j.1467-7652.2012.00732.x. Epub 2012 Aug 24. Plant Biotechnol J. 2012. PMID: 22924954
-
Identification of two 6'-deoxychalcone 4'-glucosyltransferase genes in dahlia (Dahlia variabilis).Planta. 2024 Apr 8;259(5):114. doi: 10.1007/s00425-024-04395-1. Planta. 2024. PMID: 38587670
-
Flower color modification by engineering of the flavonoid biosynthetic pathway: practical perspectives.Biosci Biotechnol Biochem. 2010;74(9):1760-9. doi: 10.1271/bbb.100358. Epub 2010 Sep 7. Biosci Biotechnol Biochem. 2010. PMID: 20834175 Review.
-
Genetic engineering of flavonoid pigments to modify flower color in floricultural plants.Biotechnol Lett. 2011 Mar;33(3):433-41. doi: 10.1007/s10529-010-0461-z. Epub 2010 Nov 4. Biotechnol Lett. 2011. PMID: 21053046 Review.
Cited by
-
Plant flavonoids--biosynthesis, transport and involvement in stress responses.Int J Mol Sci. 2013 Jul 17;14(7):14950-73. doi: 10.3390/ijms140714950. Int J Mol Sci. 2013. PMID: 23867610 Free PMC article. Review.
-
Comparative Metabolomic Analysis of the Nutritional Aspects from Ten Cultivars of the Strawberry Fruit.Foods. 2023 Mar 9;12(6):1153. doi: 10.3390/foods12061153. Foods. 2023. PMID: 36981080 Free PMC article.
-
Transcriptomic and Metabolomic Insights into Key Genes Involved in Kinsenoside Biosynthesis in Anoectochilus roxburghii.Plants (Basel). 2025 Feb 24;14(5):688. doi: 10.3390/plants14050688. Plants (Basel). 2025. PMID: 40094578 Free PMC article.
-
Six Uridine-Diphosphate Glycosyltransferases Catalyze the Glycosylation of Bioactive C13-Apocarotenols.Plant Physiol. 2020 Dec;184(4):1744-1761. doi: 10.1104/pp.20.00953. Epub 2020 Oct 5. Plant Physiol. 2020. PMID: 33020252 Free PMC article.
-
Frontiers of torenia research: innovative ornamental traits and study of ecological interaction networks through genetic engineering.Plant Methods. 2013 Jun 26;9(1):23. doi: 10.1186/1746-4811-9-23. Plant Methods. 2013. PMID: 23803155 Free PMC article.
References
-
- Tanaka Y., Katsumoto Y., Brugliera F., Mason J. Plant Cell Tissue Organ Cult. 2005;80:1–24.
-
- Harborne J. B., Baxter H. The Handbook of Natural Flavonoids 2. London: Wiley; 1999. pp. 196–201.
-
- Davies K. M., Bloor S. J., Spiller G. B., Deroles S. C. Plant J. 1998;13:259–266.
-
- Schwarz-Sommer Z., Davies B., Hudson A. Nat. Rev. Genet. 2003;4:655–664. - PubMed
-
- Asen S., Norris K. H., Stewart R. N. Phytochemistry. 1972;11:2739–2741.
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

