Dynamics, energetics, and structure in protein folding
- PMID: 16834320
- PMCID: PMC2546509
- DOI: 10.1021/bi060643c
Dynamics, energetics, and structure in protein folding
Abstract
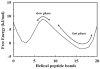
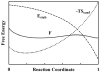
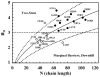
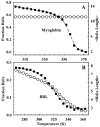
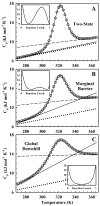
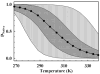
For many decades, protein folding experimentalists have worked with no information about the time scales of relevant protein folding motions and without methods for estimating the height of folding barriers. Protein folding experiments have been interpreted using chemical models in which the folding process is characterized as a series of equilibria between two or more distinct states that interconvert with activated kinetics. Accordingly, the information to be extracted from experiments was circumscribed to apparent equilibrium constants and relative folding rates. Recent developments are changing this situation dramatically. The combination of fast-folding experiments with the development of analytical methods more closely connected to physical theory reveals that folding barriers in native conditions range from minimally high (approximately 14RT for the very slow folder AcP) to nonexistent. While slow-folding (i.e., > or = 1 ms) single-domain proteins are expected to fold in a two-state fashion, microsecond-folding proteins should exhibit complex behavior arising from crossing marginal or negligible folding barriers. This realization opens a realm of exciting opportunities for experimentalists. The free energy surface of a protein with a marginal (or no) barrier can be mapped using equilibrium experiments, which could resolve energetic factors from structural factors in folding. Kinetic experiments on these proteins provide the unique opportunity to measure folding dynamics directly. Furthermore, the complex distributions of time-dependent folding behaviors expected for these proteins might be accessible to single-molecule measurements. Here, we discuss some of these recent developments in protein folding, emphasizing aspects that can serve as a guide for experimentalists interested in exploiting this new avenue of research.
Figures

References
-
- Berne BJ, Borkovec M, Straub JE. Classical and Modern Methods in Reaction-Rate Theory. J Phys Chem. 1988;92:3711–3725.
-
- Hanggi P, Talkner P, Borkovec M. Reaction-rate theory: fifty years after Kramers. Rev Mod Phys. 1990;62:251–341.
-
- Bryngelson JD, Onuchic JN, Socci ND, Wolynes PG. Funnels, Pathways, and the Energy Landscape of Protein-Folding - a Synthesis. Proteins: Struct Funct, Genet. 1995;21:167–195. - PubMed
-
- Onuchic JN, LutheySchulten Z, Wolynes PG. Theory of protein folding: The energy landscape perspective. Ann Rev Phys Chem. 1997;48:545–600. - PubMed
-
- Daggett V, Levitt M. Realistic Simulations of Native-Protein Dynamics in Solution and Beyond. Ann Rev Biophys Biomol Struct. 1993;22:353–380. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

