Programmable oligomers for minor groove DNA recognition
- PMID: 16834381
- PMCID: PMC2547997
- DOI: 10.1021/ja0621795
Programmable oligomers for minor groove DNA recognition
Abstract
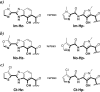
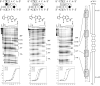
The four Watson-Crick base pairs of DNA can be distinguished in the minor groove by pairing side-by-side three five-membered aromatic carboxamides, imidazole (Im), pyrrole (Py), and hydroxypyrrole (Hp), four different ways. On the basis of the paradigm of unsymmetrical paired edges of aromatic rings for minor groove recognition, a second generation set of heterocycle pairs, imidazopyridine/pyrrole (Ip/Py) and hydroxybenzimidazole/pyrrole (Hz/Py), revealed that recognition elements not based on analogues of distamycin could be realized. A new set of end-cap heterocycle dimers, oxazole-hydroxybenzimidazole (No-Hz) and chlorothiophene-hydroxybenzimidazole (Ct-Hz), paired with Py-Py are shown to bind contiguous base pairs of DNA in the minor groove, specifically 5'-GT-3' and 5'-TT-3', with high affinity and selectivity. Utilizing this technology, we have developed a new class of oligomers for sequence-specific DNA minor groove recognition no longer based on the N-methyl pyrrole carboxamides of distamycin.
Figures



References
-
- Finlay AC, Hochstein FA, Sobin BA, Murphy FX. J. Am. Chem. Soc. 1951;73:341–343.
- Arcamone FNV, Penco S, Orezzi P, Nicolella V, Pirelli A. Nature. 1964;203:1064–1065. - PubMed
-
- Kopka ML, Yoon C, Goodsell D, Pjura P, Dickerson RE. Proc. Natl. Acad. Sci. U.S.A. 1985;82:1376–1380. - PMC - PubMed
- Pelton JG, Wemmer DE. Proc. Natl. Acad. Sci. U.S.A. 1989;86:5723–5727. - PMC - PubMed
- Wemmer DE. Annu. Rev. Biomol. Struct. 2000;29:439–461. - PubMed
- Buchmueller KL, Staples AM, Howard CM, Horick SM, Uthe PB, Minh Le N, Cox KK, Nguyen B, Pacheco KAO, Wilson DW, Lee M. J. Am. Chem. Soc. 2004;127:742–750. - PubMed
- Baraldi PG, Bovero A, Fruttarolo F, Preti D, Tabrizi MA, Pavani MG, Romagnoli R. Med. Res. Rev. 2004;24:475–528. - PubMed
- Reddy PM, Toporowski JW, Kahane AL, Bruice TC. Bioorg. Med. Chem. Lett. 2005;15:5531–5536. - PubMed
-
- White S, Szewczyk JW, Turner JM, Baird EE, Dervan PB. Nature. 1998;391:468–471. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

