Phosphorylation status of the Kep1 protein alters its affinity for its protein binding partner alternative splicing factor ASF/SF2
- PMID: 16834570
- PMCID: PMC1635453
- DOI: 10.1042/BJ20060384
Phosphorylation status of the Kep1 protein alters its affinity for its protein binding partner alternative splicing factor ASF/SF2
Abstract
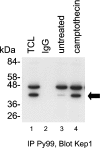
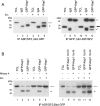
Mutations in the Drosophila kep1 gene, encoding a single maxi KH (K homology) domain-containing RNA-binding protein, result in a reduction of fertility in part due to the disruption of the apoptotic programme during oogenesis. This disruption is concomitant with the appearance of an alternatively spliced mRNA isoform encoding the inactive caspase dredd. We generated a Kep1 antibody and have found that the Kep1 protein is present in the nuclei of both the follicle and nurse cells during all stages of Drosophila oogenesis. We have shown that the Kep1 protein is phosphorylated in ovaries induced to undergo apoptosis following treatment with the topoisomerase I inhibitor camptothecin. We have also found that the Kep1 protein interacts specifically with the SR (serine/arginine-rich) protein family member ASF/SF2 (alternative splicing factor/splicing factor 2). This interaction is independent of the ability of Kep1 to bind RNA, but is dependent on the phosphorylation of the Kep1 protein, with the interaction between Kep1 and ASF/SF2 increasing in the presence of activated Src. Using a CD44v5 alternative splicing reporter construct, we observed 99% inclusion of the alternatively spliced exon 5 following kep1 transfection in a cell line that constitutively expresses activated Src. This modulation in splicing was not observed in the parental NIH 3T3 cell line in which we obtained 7.5% exon 5 inclusion following kep1 transfection. Our data suggest a mechanism of action in which the in vivo phosphorylation status of the Kep1 protein affects its affinity towards its protein binding partners and in turn may allow for the modulation of alternative splice site selection in Kep1-ASF/SF2-dependent target genes.
Figures

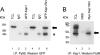


References
-
- Schwerk C., Schulze-Osthoff K. Regulation of apoptosis by alternative re-mRNA splicing. Mol. Cell. 2005;19:1–13. - PubMed
-
- Wang L., Miura M., Bergeron L., Zhu H., Yuan J. Ich-1, an Ice/ced-3-related gene, encodes both positive and negative regulators of programmed cell death. Cell. 1994;78:739–750. - PubMed
-
- Boise L. H., Gonzalez-Garcia M., Postema C. E., Ding L., Lindsten T., Turka L. A., Mao X., Nunez G., Thompson C. B. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell. 1993;74:597–608. - PubMed
-
- Shaham S., Horvitz H. R. An alternatively spliced C. elegans ced-4 RNA encodes a novel cell death inhibitor. Cell. 1996;86:201–208. - PubMed
-
- Srinivasula S. M., Ahmad M., Guo Y., Zhan Y., Lazebnik Y., Fernandes-Alnemri T., Alnemri E. S. Identification of an endogenous dominant-negative short isoform of caspase-9 that can regulate apoptosis. Cancer Res. 1999;59:999–1002. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

