Structure of UDP-N-acetylglucosamine acyltransferase with a bound antibacterial pentadecapeptide
- PMID: 16835299
- PMCID: PMC1544142
- DOI: 10.1073/pnas.0604465103
Structure of UDP-N-acetylglucosamine acyltransferase with a bound antibacterial pentadecapeptide
Abstract
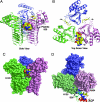
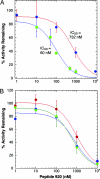
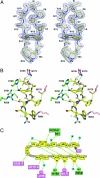
UDP-GlcNAc acyltransferase (LpxA) catalyzes the first step of lipid A biosynthesis, the transfer of the R-3-hydroxyacyl chain from R-3-hydroxyacyl acyl carrier protein (ACP) to the glucosamine 3-OH group of UDP-GlcNAc. LpxA is essential for the growth of Escherichia coli and related Gram-negative bacteria. The crystal structure of the E. coli LpxA homotrimer, determined previously at 2.6 A in the absence of substrates or inhibitors, revealed that LpxA contains an unusual, left-handed parallel beta-helix fold. We now present the crystal structure at 1.8 A resolution of E. coli LpxA in a complex with a pentadecapeptide, peptide 920. Three peptides, each of which adopts a beta-hairpin conformation, are bound per LpxA trimer. The peptides are located at the interfaces of adjacent subunits in the vicinity of the three active sites. Each peptide interacts with residues from both adjacent subunits. Peptide 920 is a potent inhibitor of E. coli LpxA (Ki = 50 nM). It is competitive with respect to acyl-ACP but not UDP-GlcNAc. The compact beta-turn structure of peptide 920 bound to LpxA may open previously uncharacterized approaches to the rational design of LpxA inhibitors with antibiotic activity.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures
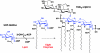

Similar articles
-
Structural basis for the acyl chain selectivity and mechanism of UDP-N-acetylglucosamine acyltransferase.Proc Natl Acad Sci U S A. 2007 Aug 21;104(34):13543-50. doi: 10.1073/pnas.0705833104. Epub 2007 Aug 13. Proc Natl Acad Sci U S A. 2007. PMID: 17698807 Free PMC article.
-
Crystal structure and activity of Francisella novicida UDP-N-acetylglucosamine acyltransferase.Biochem Biophys Res Commun. 2016 Sep 23;478(3):1223-9. doi: 10.1016/j.bbrc.2016.08.098. Epub 2016 Aug 19. Biochem Biophys Res Commun. 2016. PMID: 27545601 Free PMC article.
-
Structure guided design of an antibacterial peptide that targets UDP-N-acetylglucosamine acyltransferase.Sci Rep. 2019 Mar 8;9(1):3947. doi: 10.1038/s41598-019-40418-8. Sci Rep. 2019. PMID: 30850651 Free PMC article.
-
Lipid a biosynthesis of multidrug-resistant pathogens - a novel drug target.Curr Pharm Des. 2013;19(36):6534-50. doi: 10.2174/13816128113199990494. Curr Pharm Des. 2013. PMID: 23829374 Review.
-
Antibacterial Drug Discovery Targeting the Lipopolysaccharide Biosynthetic Enzyme LpxC.Cold Spring Harb Perspect Med. 2016 Jul 1;6(7):a025304. doi: 10.1101/cshperspect.a025304. Cold Spring Harb Perspect Med. 2016. PMID: 27235477 Free PMC article. Review.
Cited by
-
Crystal structure of LpxK, the 4'-kinase of lipid A biosynthesis and atypical P-loop kinase functioning at the membrane interface.Proc Natl Acad Sci U S A. 2012 Aug 7;109(32):12956-61. doi: 10.1073/pnas.1206072109. Epub 2012 Jul 23. Proc Natl Acad Sci U S A. 2012. PMID: 22826246 Free PMC article.
-
Targeting LPS biosynthesis and transport in gram-negative bacteria in the era of multi-drug resistance.Biochim Biophys Acta Mol Cell Res. 2023 Mar;1870(3):119407. doi: 10.1016/j.bbamcr.2022.119407. Epub 2022 Dec 18. Biochim Biophys Acta Mol Cell Res. 2023. PMID: 36543281 Free PMC article. Review.
-
Lipid A modification systems in gram-negative bacteria.Annu Rev Biochem. 2007;76:295-329. doi: 10.1146/annurev.biochem.76.010307.145803. Annu Rev Biochem. 2007. PMID: 17362200 Free PMC article. Review.
-
Discovery of dual-activity small-molecule ligands of Pseudomonas aeruginosa LpxA and LpxD using SPR and X-ray crystallography.Sci Rep. 2019 Oct 29;9(1):15450. doi: 10.1038/s41598-019-51844-z. Sci Rep. 2019. PMID: 31664082 Free PMC article.
-
A Rab-centric perspective of bacterial pathogen-occupied vacuoles.Cell Host Microbe. 2013 Sep 11;14(3):256-68. doi: 10.1016/j.chom.2013.08.010. Cell Host Microbe. 2013. PMID: 24034612 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

