Sequential processing of a mitochondrial tandem protein: insights into protein import in Schizosaccharomyces pombe
- PMID: 16835444
- PMCID: PMC1489288
- DOI: 10.1128/EC.00092-06
Sequential processing of a mitochondrial tandem protein: insights into protein import in Schizosaccharomyces pombe
Abstract
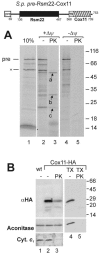
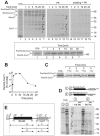
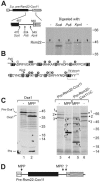
The sequencing of the genome of Schizosaccharomyces pombe revealed the presence of a number of genes encoding tandem proteins, some of which are mitochondrial components. One of these proteins (pre-Rsm22-Cox11) consists of a fusion of Rsm22, a component of the mitochondrial ribosome, and Cox11, a factor required for copper insertion into cytochrome oxidase. Since in Saccharomyces cerevisiae, Cox11 is physically attached to the mitochondrial ribosome, it was suggested that the tandem organization of Rsm22-Cox11 is used to covalently tie the mitochondrial ribosome to Cox11 in S. pombe. We report here that pre-Rsm22-Cox11 is matured in two subsequent processing events. First, the mitochondrial presequence is removed. At a later stage of the import process, the Rsm22 and Cox11 domains are separated by cleavage of the mitochondrial processing peptidase at an internal processing site. In vivo data obtained using a tagged version of pre-Rsm22-Cox11 confirmed the proteolytic separation of Cox11 from the Rsm22 domain. Hence, the tandem organization of pre-Rsm22-Cox11 does not give rise to a persistent fusion protein but rather might be used to increase the import efficiency of Cox11 and/or to coordinate expression levels of Rsm22 and Cox11 in S. pombe.
Figures



Similar articles
-
Functional analysis of the domains in Cox11.J Biol Chem. 2005 Jun 17;280(24):22664-9. doi: 10.1074/jbc.M414077200. Epub 2005 Apr 19. J Biol Chem. 2005. PMID: 15840584
-
The fission yeast Schizosaccharomyces pombe has two distinct tRNase Z(L)s encoded by two different genes and differentially targeted to the nucleus and mitochondria.Biochem J. 2011 Apr 1;435(1):103-11. doi: 10.1042/BJ20101619. Biochem J. 2011. PMID: 21208191
-
Redox-regulated dynamic interplay between Cox19 and the copper-binding protein Cox11 in the intermembrane space of mitochondria facilitates biogenesis of cytochrome c oxidase.Mol Biol Cell. 2015 Jul 1;26(13):2385-401. doi: 10.1091/mbc.E14-11-1526. Epub 2015 Apr 29. Mol Biol Cell. 2015. PMID: 25926683 Free PMC article.
-
Protein import into mitochondria.IUBMB Life. 2001 Sep-Nov;52(3-5):101-12. doi: 10.1080/15216540152845894. IUBMB Life. 2001. PMID: 11798021 Review.
-
Protein insertion into the inner membrane of mitochondria.IUBMB Life. 2003 Apr-May;55(4-5):219-25. doi: 10.1080/1521654031000123349. IUBMB Life. 2003. PMID: 12880202 Review.
Cited by
-
The Assembly Factor Pet117 Couples Heme a Synthase Activity to Cytochrome Oxidase Assembly.J Biol Chem. 2017 Feb 3;292(5):1815-1825. doi: 10.1074/jbc.M116.766980. Epub 2016 Dec 20. J Biol Chem. 2017. PMID: 27998984 Free PMC article.
-
More than just a ticket canceller: the mitochondrial processing peptidase tailors complex precursor proteins at internal cleavage sites.Mol Biol Cell. 2020 Nov 15;31(24):2657-2668. doi: 10.1091/mbc.E20-08-0524. Epub 2020 Sep 30. Mol Biol Cell. 2020. PMID: 32997570 Free PMC article.
-
Proteomic profiling of the mitochondrial ribosome identifies Atp25 as a composite mitochondrial precursor protein.Mol Biol Cell. 2016 Oct 15;27(20):3031-3039. doi: 10.1091/mbc.E16-07-0513. Epub 2016 Aug 31. Mol Biol Cell. 2016. PMID: 27582385 Free PMC article.
-
The UPRmt preserves mitochondrial import to extend lifespan.J Cell Biol. 2022 Jul 4;221(7):e202201071. doi: 10.1083/jcb.202201071. Epub 2022 May 24. J Cell Biol. 2022. PMID: 35608535 Free PMC article.
-
Mne1 is a novel component of the mitochondrial splicing apparatus responsible for processing of a COX1 group I intron in yeast.J Biol Chem. 2011 Mar 25;286(12):10137-46. doi: 10.1074/jbc.M110.205625. Epub 2011 Jan 21. J Biol Chem. 2011. PMID: 21257754 Free PMC article.
References
-
- Albury, M. S., P. Dudley, F. Z. Watts, and A. L. Moore. 1996. Targeting the plant alternative oxidase protein to Schizosaccharomyces pombe mitochondria confers cyanide-insensitive respiration. J. Biol. Chem. 271:17062-17066. - PubMed
-
- Barros, M. H., C. G. Carlson, D. M. Glerum, and A. Tzagoloff. 2001. Involvement of mitochondrial ferredoxin and Cox15p in hydroxylation of heme O. FEBS Lett. 492:133-138. - PubMed
-
- Bureik, M., B. Schiffler, Y. Hiraoka, F. Vogel, and R. Bernhardt. 2002. Functional expression of human mitochondrial CYP11B2 in fission yeast and identification of a new internal electron transfer protein, etp1. Biochemistry 41:2311-2321. - PubMed
-
- Carr, H. S., A. B. Maxfield, Y. C. Horng, and D. R. Winge. 2005. Functional analysis of the domains in Cox11. J. Biol. Chem. 280:22664-22669. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

