A MADS box protein interacts with a mating-type protein and is required for fruiting body development in the homothallic ascomycete Sordaria macrospora
- PMID: 16835449
- PMCID: PMC1489284
- DOI: 10.1128/EC.00086-06
A MADS box protein interacts with a mating-type protein and is required for fruiting body development in the homothallic ascomycete Sordaria macrospora
Abstract
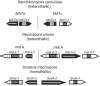
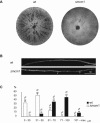
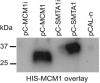
MADS box transcription factors control diverse developmental processes in plants, metazoans, and fungi. To analyze the involvement of MADS box proteins in fruiting body development of filamentous ascomycetes, we isolated the mcm1 gene from the homothallic ascomycete Sordaria macrospora, which encodes a putative homologue of the Saccharomyces cerevisiae MADS box protein Mcm1p. Deletion of the S. macrospora mcm1 gene resulted in reduced biomass, increased hyphal branching, and reduced hyphal compartment length during vegetative growth. Furthermore, the S. macrospora Deltamcm1 strain was unable to produce fruiting bodies or ascospores during sexual development. A yeast two-hybrid analysis in conjugation with in vitro analyses demonstrated that the S. macrospora MCM1 protein can interact with the putative transcription factor SMTA-1, encoded by the S. macrospora mating-type locus. These results suggest that the S. macrospora MCM1 protein is involved in the transcriptional regulation of mating-type-specific genes as well as in fruiting body development.
Figures






Similar articles
-
Autophagy genes Smatg8 and Smatg4 are required for fruiting-body development, vegetative growth and ascospore germination in the filamentous ascomycete Sordaria macrospora.Autophagy. 2013 Jan;9(1):33-49. doi: 10.4161/auto.22398. Epub 2012 Oct 12. Autophagy. 2013. PMID: 23064313 Free PMC article.
-
A STE12 homologue of the homothallic ascomycete Sordaria macrospora interacts with the MADS box protein MCM1 and is required for ascosporogenesis.Mol Microbiol. 2006 Nov;62(3):853-68. doi: 10.1111/j.1365-2958.2006.05415.x. Epub 2006 Sep 25. Mol Microbiol. 2006. PMID: 16999832
-
Autophagy-Associated Protein SmATG12 Is Required for Fruiting-Body Formation in the Filamentous Ascomycete Sordaria macrospora.PLoS One. 2016 Jun 16;11(6):e0157960. doi: 10.1371/journal.pone.0157960. eCollection 2016. PLoS One. 2016. PMID: 27309377 Free PMC article.
-
Sordaria macrospora: 25 years as a model organism for studying the molecular mechanisms of fruiting body development.Appl Microbiol Biotechnol. 2020 May;104(9):3691-3704. doi: 10.1007/s00253-020-10504-3. Epub 2020 Mar 11. Appl Microbiol Biotechnol. 2020. PMID: 32162092 Free PMC article. Review.
-
The filamentous fungus Sordaria macrospora as a genetic model to study fruiting body development.Adv Genet. 2014;87:199-244. doi: 10.1016/B978-0-12-800149-3.00004-4. Adv Genet. 2014. PMID: 25311923 Review.
Cited by
-
Autophagy genes Smatg8 and Smatg4 are required for fruiting-body development, vegetative growth and ascospore germination in the filamentous ascomycete Sordaria macrospora.Autophagy. 2013 Jan;9(1):33-49. doi: 10.4161/auto.22398. Epub 2012 Oct 12. Autophagy. 2013. PMID: 23064313 Free PMC article.
-
MADS-box transcription factor mig1 is required for infectious growth in Magnaporthe grisea.Eukaryot Cell. 2008 May;7(5):791-9. doi: 10.1128/EC.00009-08. Epub 2008 Mar 14. Eukaryot Cell. 2008. PMID: 18344407 Free PMC article.
-
Functional characterization of MAT1-1-specific mating-type genes in the homothallic ascomycete Sordaria macrospora provides new insights into essential and nonessential sexual regulators.Eukaryot Cell. 2010 Jun;9(6):894-905. doi: 10.1128/EC.00019-10. Epub 2010 Apr 30. Eukaryot Cell. 2010. PMID: 20435701 Free PMC article.
-
Molecular mechanisms underlying phenotypic degeneration in Cordyceps militaris: insights from transcriptome reanalysis and osmotic stress studies.Sci Rep. 2024 Jan 26;14(1):2231. doi: 10.1038/s41598-024-51946-3. Sci Rep. 2024. PMID: 38278834 Free PMC article.
-
The phocein homologue SmMOB3 is essential for vegetative cell fusion and sexual development in the filamentous ascomycete Sordaria macrospora.Curr Genet. 2011 Apr;57(2):133-49. doi: 10.1007/s00294-010-0333-z. Epub 2011 Jan 13. Curr Genet. 2011. PMID: 21229248 Free PMC article.
References
-
- Alvarez-Buylla, E. R., S. Pelaz, S. J. Liljegren, S. E. Gold, C. Burgeff, G. S. Ditta, L. Ribas de Pouplana, L. Martinez-Castilla, and M. F. Yanofsky. 2000. An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. USA 97:5328-5333. - PMC - PubMed
-
- Becker, A., and G. Theissen. 2003. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet. Evol. 29:464-489. - PubMed
-
- Becker, D. M., and V. Lundblad. 1994. Introduction of DNA into yeast cells, p. 13.7.1-13.7.10. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. Wiley, New York, N.Y. - PubMed
-
- Bender, A., and G. F. Sprague, Jr. 1987. MAT alpha 1 protein, a yeast transcription activator, binds synergistically with a second protein to a set of cell-type-specific genes. Cell 50:681-691. - PubMed
-
- Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

