Mapping of glycolytic enzyme-binding sites on human erythrocyte band 3
- PMID: 16836485
- PMCID: PMC1635443
- DOI: 10.1042/BJ20060792
Mapping of glycolytic enzyme-binding sites on human erythrocyte band 3
Abstract
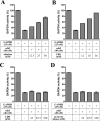
Previous work has shown that GAPDH (glyceraldehyde-3-phosphate dehydrogenase), aldolase, PFK (phosphofructokinase), PK (pyruvate kinase) and LDH (lactate dehydrogenase) assemble into a GE (glycolytic enzyme) complex on the inner surface of the human erythrocyte membrane. In an effort to define the molecular architecture of this complex, we have undertaken to localize the binding sites of these enzymes more accurately. We report that: (i) a major aldolase-binding site on the erythrocyte membrane is located within N-terminal residues 1-23 of band 3 and that both consensus sequences D6DYED10 and E19EYED23 are necessary to form a single enzyme-binding site; (ii) GAPDH has two tandem binding sites on band 3, located in residues 1-11 and residues 12-23 respectively; (iii) a PFK-binding site resides between residues 12 and 23 of band 3; (iv) no GEs bind to the third consensus sequence (residues D902EYDE906) at the C-terminus of band 3; and (v) the LDH- and PK-binding sites on the erythrocyte membrane do not reside on band 3. Taken together, these results argue that band 3 provides a nucleation site for the GE complex on the human erythrocyte membrane and that other components near band 3 must also participate in organizing the enzyme complex.
Figures




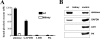

References
-
- Hoffman J. F. ATP compartmentation in human erythrocytes. Curr. Opin. Hematol. 1997;4:112–115. - PubMed
-
- Strapazon E., Steck T. L. Interaction of the aldolase and the membrane of human erythrocytes. Biochemistry. 1977;16:2966–2971. - PubMed
-
- Murthy S. N., Liu T., Kaul R. K., Kohler H., Steck T. L. The aldolase-binding site of the human erythrocyte membrane is at the NH2 terminus of band 3. J. Biol. Chem. 1981;256:11203–11208. - PubMed
-
- Tsai I. H., Murthy S. N., Steck T. L. Effect of red cell membrane binding on the catalytic activity of glyceraldehyde-3-phosphate dehydrogenase. J. Biol. Chem. 1982;257:1438–1442. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

