Immune-privileged embryonic Swiss mouse STO and STO cell-derived progenitor cells: major histocompatibility complex and cell differentiation antigen expression patterns resemble those of human embryonic stem cell lines
- PMID: 16836618
- PMCID: PMC1782333
- DOI: 10.1111/j.1365-2567.2006.02412.x
Immune-privileged embryonic Swiss mouse STO and STO cell-derived progenitor cells: major histocompatibility complex and cell differentiation antigen expression patterns resemble those of human embryonic stem cell lines
Abstract
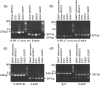
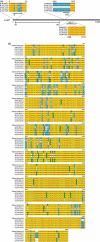
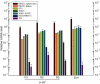
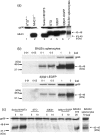
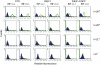
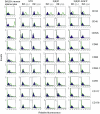
Embryonic mouse STO (S, SIM; T, 6-thioguanine resistant; O, ouabain resistant) and 3(8)21-enhanced green fluorescent protein (EGFP) cell lines exhibit long-term survival and hepatic progenitor cell behaviour after xenogeneic engraftment in non-immunosuppressed inbred rats, and were previously designated major histocompatibility complex (MHC) class I- and class II-negative lines. To determine the molecular basis for undetectable MHC determinants, the expression and haplotype of H-2K, H-2D, H-2L and I-A proteins were reassessed by reverse transcriptase-polymerase chain reaction (RT-PCR), cDNA sequencing, RNA hybridization, immunoblotting, quantitative RT-PCR (QPCR), immunocytochemistry and flow cytometry. To detect cell differentiation (CD) surface antigens characteristic of stem cells, apoptotic regulation or adaptive immunity that might facilitate progenitor cell status or immune privilege, flow cytometry was also used to screen untreated and cytokine [interferon (IFN)-gamma]-treated cultures. Despite prior PCR genotyping analyses suggestive of H-2q haplotypes in STO, 3(8)21-EGFP and parental 3(8)21 cells, all three lines expressed H-2K cDNA sequences identical to those of d-haplotype BALB/c mice, as well as constitutive and cytokine-inducible H-2K(d) determinants. In contrast, apart from H-2L(d[LOW]) display in 3(8)21 cells, H-2Dd, H-2Ld and I-Ad determinants were undetectable. All three lines expressed constitutive and cytokine-inducible CD34; however, except for inducible CD117([LOW]) expression in 3(8)21 cells, no expression of CD45, CD117, CD62L, CD80, CD86, CD90.1 or CD95L/CD178 was observed. Constitutive and cytokine-inducible CD95([LOW]) expression was detected in STO and 3(8)21 cells, but not in 3(8)21-EGFP cells. MHC (class I(+[LOW])/class II-) and CD (CD34+/CD80-/CD86-/CD95L-) expression patterns in STO and STO cell-derived progenitor cells resemble patterns reported for human embryonic stem cell lines. Whether these patterns reflect associations with mechanisms that are regulatory of immune privilege or functional tissue-specific plasticity is unknown.
Figures
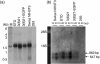


Similar articles
-
Hepatic progenitor cell lines from allyl alcohol-treated adult rats are derived from gamma-irradiated mouse STO cells.Stem Cells. 2003;21(4):449-58. doi: 10.1634/stemcells.21-4-449. Stem Cells. 2003. PMID: 12832698
-
Nucleotide sequences of three H-2K and three H-2D complementary DNA clones coding mouse class I MHC heavy chain proteins.Ann Transplant. 1996;1(3):26-31. Ann Transplant. 1996. PMID: 9869916
-
Activation of antigen-specific cytotoxic T lymphocytes by beta 2-microglobulin or TAP1 gene disruption and the introduction of recipient-matched MHC class I gene in allogeneic embryonic stem cell-derived dendritic cells.J Immunol. 2008 Nov 1;181(9):6635-43. doi: 10.4049/jimmunol.181.9.6635. J Immunol. 2008. PMID: 18941254
-
Major histocompatibility complex regulation of cytokine production.J Interferon Cytokine Res. 1996 Dec;16(12):983-8. doi: 10.1089/jir.1996.16.983. J Interferon Cytokine Res. 1996. PMID: 8973998 Review.
-
Inhibition of major histocompatibility complex class I antigen shedding up-regulates the surface expression of class I antigens on the lymphocyte cell surface.Ann N Y Acad Sci. 1991;628:354-61. doi: 10.1111/j.1749-6632.1991.tb17268.x. Ann N Y Acad Sci. 1991. PMID: 1906254 Review.
Cited by
-
Human embryonic stem cell-extracts inhibit the differentiation and function of monocyte-derived dendritic cells.Stem Cell Rev Rep. 2010 Dec;6(4):611-21. doi: 10.1007/s12015-010-9185-7. Stem Cell Rev Rep. 2010. PMID: 20711689
-
Embryonic stem cell-derived factors inhibit T effector activation and induce T regulatory cells by suppressing PKC-θ activation.PLoS One. 2012;7(3):e32420. doi: 10.1371/journal.pone.0032420. Epub 2012 Mar 7. PLoS One. 2012. PMID: 22412871 Free PMC article.
-
Targeted deletion of hepatocyte Ikkbeta confers growth advantages.Biochem Biophys Res Commun. 2009 Mar 6;380(2):349-54. doi: 10.1016/j.bbrc.2009.01.085. Epub 2009 Jan 24. Biochem Biophys Res Commun. 2009. PMID: 19171122 Free PMC article.
-
Dendritic cell-based immunotherapy in prevention and treatment of renal cell carcinoma: efficacy, safety, and activity of Ad-GM·CAIX in immunocompetent mouse models.J Immunother. 2013 Feb;36(2):102-11. doi: 10.1097/CJI.0b013e31827bec97. J Immunother. 2013. PMID: 23377663 Free PMC article.
-
The immune boundaries for stem cell based therapies: problems and prospective solutions.J Cell Mol Med. 2009 Aug;13(8A):1464-75. doi: 10.1111/j.1582-4934.2009.00837.x. Epub 2009 Jul 6. J Cell Mol Med. 2009. PMID: 19583810 Free PMC article. Review.
References
-
- Auchincloss H., Jr In search of the elusive Holy Grail: the mechanisms and prospects for achieving clinical transplantation tolerance. Am J Transplant. 2001;1:6–12. - PubMed
-
- Kamradt T, Mitchison NA. Tolerance and autoimmunity. N Engl J Med. 2001;344:655–64. - PubMed
-
- Bellingham RE, Brent L, Medawar PB. ‘Actively acquired tolerance’ of foreign cells. Nature. 1953;172:603–6. - PubMed
-
- Liechty KW, MacKenzie TC, Shaaban AF, Radu A, Mosely AM, Deans R, Marshak DR, Flake AW. Human mesenchymal stem cells engraft and demonstrate site-specific differentiation after in utero transplantation in sheep. Nature Med. 2000;6:1282–6. - PubMed
-
- Taylor AW, Streilein JW. Inhibition of antigen-stimulated effector T cells by human cerebrospinal fluid. Neuroimmunomodulation. 1996;3:112–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

