The claudin gene family: expression in normal and neoplastic tissues
- PMID: 16836752
- PMCID: PMC1538620
- DOI: 10.1186/1471-2407-6-186
The claudin gene family: expression in normal and neoplastic tissues
Abstract
Background: The claudin (CLDN) genes encode a family of proteins important in tight junction formation and function. Recently, it has become apparent that CLDN gene expression is frequently altered in several human cancers. However, the exact patterns of CLDN expression in various cancers is unknown, as only a limited number of CLDN genes have been investigated in a few tumors.
Methods: We identified all the human CLDN genes from Genbank and we used the large public SAGE database to ascertain the gene expression of all 21 CLDN in 266 normal and neoplastic tissues. Using real-time RT-PCR, we also surveyed a subset of 13 CLDN genes in 24 normal and 24 neoplastic tissues.
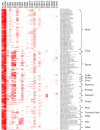
Results: We show that claudins represent a family of highly related proteins, with claudin-16, and -23 being the most different from the others. From in silico analysis and RT-PCR data, we find that most claudin genes appear decreased in cancer, while CLDN3, CLDN4, and CLDN7 are elevated in several malignancies such as those originating from the pancreas, bladder, thyroid, fallopian tubes, ovary, stomach, colon, breast, uterus, and the prostate. Interestingly, CLDN5 is highly expressed in vascular endothelial cells, providing a possible target for antiangiogenic therapy. CLDN18 might represent a biomarker for gastric cancer.
Conclusion: Our study confirms previously known CLDN gene expression patterns and identifies new ones, which may have applications in the detection, prognosis and therapy of several human cancers. In particular we identify several malignancies that express CLDN3 and CLDN4. These cancers may represent ideal candidates for a novel therapy being developed based on CPE, a toxin that specifically binds claudin-3 and claudin-4.
Figures


Similar articles
-
Claudin expression in breast cancer: high or low, what to expect?Histol Histopathol. 2012 Oct;27(10):1283-95. doi: 10.14670/HH-27.1283. Histol Histopathol. 2012. PMID: 22936447
-
CLDN23 gene, frequently down-regulated in intestinal-type gastric cancer, is a novel member of CLAUDIN gene family.Int J Mol Med. 2003 Jun;11(6):683-9. Int J Mol Med. 2003. PMID: 12736707
-
[Claudin expression in different pancreatic cancers and its significance in differential diagnostics].Magy Onkol. 2009 Sep;53(3):273-8. doi: 10.1556/MOnkol.53.2009.3.7. Magy Onkol. 2009. PMID: 19793693 Hungarian.
-
Claudin-1, A Double-Edged Sword in Cancer.Int J Mol Sci. 2020 Jan 15;21(2):569. doi: 10.3390/ijms21020569. Int J Mol Sci. 2020. PMID: 31952355 Free PMC article. Review.
-
Roles of the first-generation claudin binder, Clostridium perfringens enterotoxin, in the diagnosis and claudin-targeted treatment of epithelium-derived cancers.Pflugers Arch. 2017 Jan;469(1):45-53. doi: 10.1007/s00424-016-1878-6. Epub 2016 Sep 15. Pflugers Arch. 2017. PMID: 27629072 Review.
Cited by
-
Molecular resistance fingerprint of pemetrexed and platinum in a long-term survivor of mesothelioma.PLoS One. 2012;7(8):e40521. doi: 10.1371/journal.pone.0040521. Epub 2012 Aug 8. PLoS One. 2012. PMID: 22905093 Free PMC article.
-
Claudin expression in pancreatic endocrine tumors as compared with ductal adenocarcinomas.Virchows Arch. 2007 May;450(5):549-57. doi: 10.1007/s00428-007-0406-7. Epub 2007 Apr 12. Virchows Arch. 2007. PMID: 17429687
-
Effects of 1,2-Dimethylhydrazine on Barrier Properties of Rat Large Intestine and IPEC-J2 Cells.Int J Mol Sci. 2021 Sep 24;22(19):10278. doi: 10.3390/ijms221910278. Int J Mol Sci. 2021. PMID: 34638619 Free PMC article.
-
Normalization of proliferation and tight junction formation in bladder epithelial cells from patients with interstitial cystitis/painful bladder syndrome by d-proline and d-pipecolic acid derivatives of antiproliferative factor.Chem Biol Drug Des. 2011 Jun;77(6):421-30. doi: 10.1111/j.1747-0285.2011.01108.x. Epub 2011 Apr 27. Chem Biol Drug Des. 2011. PMID: 21352500 Free PMC article.
-
CLDN10 promotes a malignant phenotype of osteosarcoma cells via JAK1/Stat1 signaling.J Cell Commun Signal. 2019 Sep;13(3):395-405. doi: 10.1007/s12079-019-00509-7. Epub 2019 Feb 22. J Cell Commun Signal. 2019. PMID: 30796717 Free PMC article.
References
-
- Hadj-Rabia S, Baala L, Vabres P, Hamel-Teillac D, Jacquemin E, Fabre M, Lyonnet S, De Prost Y, Munnich A, Hadchouel M, Smahi A. Claudin-1 gene mutations in neonatal sclerosing cholangitis associated with ichthyosis: a tight junction disease. Gastroenterology. 2004;127:1386–1390. doi: 10.1053/j.gastro.2004.07.022. - DOI - PubMed
-
- Wilcox ER, Burton QL, Naz S, Riazuddin S, Smith TN, Ploplis B, Belyantseva I, Ben-Yosef T, Liburd NA, Morell RJ, Kachar B, Wu DK, Griffith AJ, Friedman TB. Mutations in the gene encoding tight junction claudin-14 cause autosomal recessive deafness DFNB29. Cell. 2001;104:165–172. doi: 10.1016/S0092-8674(01)00200-8. - DOI - PubMed
-
- Simon DB, Lu Y, Choate KA, Velazquez H, Al-Sabban E, Praga M, Casari G, Bettinelli A, Colussi G, Rodriguez-Soriano J, McCredie D, Milford D, Sanjad S, Lifton RP. Paracellin-1, a renal tight junction protein required for paracellular Mg2+ resorption. Science. 1999;285:103–106. doi: 10.1126/science.285.5424.103. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

