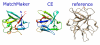Tools for integrated sequence-structure analysis with UCSF Chimera
- PMID: 16836757
- PMCID: PMC1570152
- DOI: 10.1186/1471-2105-7-339
Tools for integrated sequence-structure analysis with UCSF Chimera
Abstract
Background: Comparing related structures and viewing the structures in the context of sequence alignments are important tasks in protein structure-function research. While many programs exist for individual aspects of such work, there is a need for interactive visualization tools that: (a) provide a deep integration of sequence and structure, far beyond mapping where a sequence region falls in the structure and vice versa; (b) facilitate changing data of one type based on the other (for example, using only sequence-conserved residues to match structures, or adjusting a sequence alignment based on spatial fit); (c) can be used with a researcher's own data, including arbitrary sequence alignments and annotations, closely or distantly related sets of proteins, etc.; and (d) interoperate with each other and with a full complement of molecular graphics features. We describe enhancements to UCSF Chimera to achieve these goals.
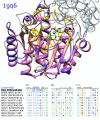
Results: The molecular graphics program UCSF Chimera includes a suite of tools for interactive analyses of sequences and structures. Structures automatically associate with sequences in imported alignments, allowing many kinds of crosstalk. A novel method is provided to superimpose structures in the absence of a pre-existing sequence alignment. The method uses both sequence and secondary structure, and can match even structures with very low sequence identity. Another tool constructs structure-based sequence alignments from superpositions of two or more proteins. Chimera is designed to be extensible, and mechanisms for incorporating user-specific data without Chimera code development are also provided.
Conclusion: The tools described here apply to many problems involving comparison and analysis of protein structures and their sequences. Chimera includes complete documentation and is intended for use by a wide range of scientists, not just those in the computational disciplines. UCSF Chimera is free for non-commercial use and is available for Microsoft Windows, Apple Mac OS X, Linux, and other platforms from http://www.cgl.ucsf.edu/chimera.
Figures