The molecular etiologies and associated phenotypes of amelogenesis imperfecta
- PMID: 16838342
- PMCID: PMC1847600
- DOI: 10.1002/ajmg.a.31358
The molecular etiologies and associated phenotypes of amelogenesis imperfecta
Abstract
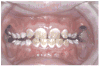
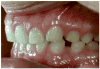
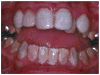
The amelogenesis imperfectas (AIs) are a clinically and genetically diverse group of conditions that are caused by mutations in a variety of genes that are critical for normal enamel formation. To date, mutations have been identified in four genes (AMELX, ENAM, KLK4, MMP20) known to be involved in enamel formation. Additional yet to be identified genes also are implicated in the etiology of AI based on linkage studies. The diverse and often unique phenotypes resulting from the different allelic and non-allelic mutations in these genes provide an opportunity to better understand the role of these genes and their related proteins in enamel formation. Understanding the AI phenotypes also provides an aid to clinicians in directing molecular studies aimed at delineating the genetic basis underlying these diverse clinical conditions. Our current knowledge of the known mutations and associated phenotypes of the different AI subtypes are reviewed.
Figures




Similar articles
-
Human and mouse enamel phenotypes resulting from mutation or altered expression of AMEL, ENAM, MMP20 and KLK4.Cells Tissues Organs. 2009;189(1-4):224-9. doi: 10.1159/000151378. Epub 2008 Aug 19. Cells Tissues Organs. 2009. PMID: 18714142 Free PMC article.
-
Exclusion of candidate genes in seven Turkish families with autosomal recessive amelogenesis imperfecta.Am J Med Genet A. 2009 Jul;149A(7):1392-8. doi: 10.1002/ajmg.a.32885. Am J Med Genet A. 2009. PMID: 19530186 Free PMC article.
-
Target gene analyses of 39 amelogenesis imperfecta kindreds.Eur J Oral Sci. 2011 Dec;119 Suppl 1(Suppl 1):311-23. doi: 10.1111/j.1600-0722.2011.00857.x. Eur J Oral Sci. 2011. PMID: 22243262 Free PMC article.
-
Relationship of phenotype and genotype in X-linked amelogenesis imperfecta.Connect Tissue Res. 2003;44 Suppl 1:72-8. Connect Tissue Res. 2003. PMID: 12952177 Review.
-
Genes and related proteins involved in amelogenesis imperfecta.J Dent Res. 2005 Dec;84(12):1117-26. doi: 10.1177/154405910508401206. J Dent Res. 2005. PMID: 16304440 Review.
Cited by
-
Enamel protein regulation and dental and periodontal physiopathology in MSX2 mutant mice.Am J Pathol. 2010 Nov;177(5):2516-26. doi: 10.2353/ajpath.2010.091224. Epub 2010 Oct 7. Am J Pathol. 2010. PMID: 20934968 Free PMC article.
-
Amelogenesis imperfecta due to a mutation of the enamelin gene: clinical case with genotype-phenotype correlations.Pediatr Dent. 2010 Jan-Feb;32(1):56-60. Pediatr Dent. 2010. PMID: 20298654 Free PMC article.
-
Tooth dentin defects reflect genetic disorders affecting bone mineralization.Bone. 2012 Apr;50(4):989-97. doi: 10.1016/j.bone.2012.01.010. Epub 2012 Jan 26. Bone. 2012. PMID: 22296718 Free PMC article. Review.
-
Possible association of amelogenin to high caries experience in a Guatemalan-Mayan population.Caries Res. 2008;42(1):8-13. doi: 10.1159/000111744. Epub 2007 Nov 27. Caries Res. 2008. PMID: 18042988 Free PMC article.
-
Weaker dental enamel explains dental decay.PLoS One. 2015 Apr 17;10(4):e0124236. doi: 10.1371/journal.pone.0124236. eCollection 2015. PLoS One. 2015. PMID: 25885796 Free PMC article.
References
-
- Aldred MJ, Crawford PJM, Roberts E, Gillespie CM, Thomas NT, Fenton I, Sandkuijl LA, Harper PS. Genetic heterogeneity in X-linked amelogenesis imperfecta. Genomics. 1992a;14:567–573. - PubMed
-
- Aldred MJ, Crawford PJM, Roberts E, Thomas NST. Identification of a nonsense mutation in the amelogenin gene (AMELX) in a family with X-linked amelogenesis imperfecta (AIH1) Hum Genet. 1992b;90:413–416. - PubMed
-
- Aldred MJ, Savarirayan R, Crawford PJM. Amelogenesis imperfecta: A classification and catalogue for the 21st century. Oral Disease. 2003;9:19–23. - PubMed
-
- Baba O, Takahashi N, Terashima T, Li W, DenBesten PK, Takano Y. Expression of alternatively spliced RNA transcripts of amelogenin gene exons 8 and 9 and its end products in the rat incisor. J Histochem Cytochem. 2002;50:1229–1236. - PubMed
-
- Backman B. Amelogenesis imperfecta-clinical manifestations in 51 families in a northern Swedish country. Scand J Dent Res. 1988;96:505–516. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

