Modeling chromosomes in mouse to explore the function of genes, genomic disorders, and chromosomal organization
- PMID: 16839184
- PMCID: PMC1500809
- DOI: 10.1371/journal.pgen.0020086
Modeling chromosomes in mouse to explore the function of genes, genomic disorders, and chromosomal organization
Abstract
One of the challenges of genomic research after the completion of the human genome project is to assign a function to all the genes and to understand their interactions and organizations. Among the various techniques, the emergence of chromosome engineering tools with the aim to manipulate large genomic regions in the mouse model offers a powerful way to accelerate the discovery of gene functions and provides more mouse models to study normal and pathological developmental processes associated with aneuploidy. The combination of gene targeting in ES cells, recombinase technology, and other techniques makes it possible to generate new chromosomes carrying specific and defined deletions, duplications, inversions, and translocations that are accelerating functional analysis. This review presents the current status of chromosome engineering techniques and discusses the different applications as well as the implication of these new techniques in future research to better understand the function of chromosomal organization and structures.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures

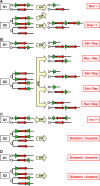

References
-
- Green EL, Roderick TH. Radiation genetics. In: Green EL, editor. Biology of the laboratory mouse. New York: McGraw-Hill; 1966. pp. 87–150.
-
- Goodwin NC, Ishida Y, Hartford S, Wnek C, Bergstrom RA, et al. DelBank: A mouse ES-cell resource for generating deletions. Nat Genet. 2001;28:310–311. - PubMed
-
- You Y, Bergstrom R, Klemm M, Lederman B, Nelson H, et al. Chromosomal deletion complexes in mice by radiation of embryonic stem cells. Nat Genet. 1997;15:285–288. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

