Neofunctionalization in vertebrates: the example of retinoic acid receptors
- PMID: 16839186
- PMCID: PMC1500811
- DOI: 10.1371/journal.pgen.0020102
Neofunctionalization in vertebrates: the example of retinoic acid receptors
Abstract
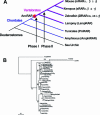
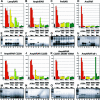
Understanding the role of gene duplications in establishing vertebrate innovations is one of the main challenges of Evo-Devo (evolution of development) studies. Data on evolutionary changes in gene expression (i.e., evolution of transcription factor-cis-regulatory elements relationships) tell only part of the story; protein function, best studied by biochemical and functional assays, can also change. In this study, we have investigated how gene duplication has affected both the expression and the ligand-binding specificity of retinoic acid receptors (RARs), which play a major role in chordate embryonic development. Mammals have three paralogous RAR genes--RAR alpha, beta, and gamma--which resulted from genome duplications at the origin of vertebrates. By using pharmacological ligands selective for specific paralogues, we have studied the ligand-binding capacities of RARs from diverse chordates species. We have found that RAR beta-like binding selectivity is a synapomorphy of all chordate RARs, including a reconstructed synthetic RAR representing the receptor present in the ancestor of chordates. Moreover, comparison of expression patterns of the cephalochordate amphioxus and the vertebrates suggests that, of all the RARs, RAR beta expression has remained most similar to that of the ancestral RAR. On the basis of these results together, we suggest that while RAR beta kept the ancestral RAR role, RAR alpha and RAR gamma diverged both in ligand-binding capacity and in expression patterns. We thus suggest that neofunctionalization occurred at both the expression and the functional levels to shape RAR roles during development in vertebrates.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures




Similar articles
-
Isotype-restricted corepressor recruitment: a constitutively closed helix 12 conformation in retinoic acid receptors beta and gamma interferes with corepressor recruitment and prevents transcriptional repression.Mol Cell Biol. 2003 Apr;23(8):2844-58. doi: 10.1128/MCB.23.8.2844-2858.2003. Mol Cell Biol. 2003. PMID: 12665583 Free PMC article.
-
Analysis of the ligand-binding domain of human retinoic acid receptor alpha by site-directed mutagenesis.Mol Cell Biol. 1996 Oct;16(10):5386-92. doi: 10.1128/MCB.16.10.5386. Mol Cell Biol. 1996. PMID: 8816450 Free PMC article.
-
Evolution of retinoic acid receptors in chordates: insights from three lamprey species, Lampetra fluviatilis, Petromyzon marinus, and Lethenteron japonicum.Evodevo. 2015 May 7;6:18. doi: 10.1186/s13227-015-0016-4. eCollection 2015. Evodevo. 2015. PMID: 25984292 Free PMC article.
-
Evolution of retinoic acid receptors and retinoic acid signaling.Subcell Biochem. 2014;70:55-73. doi: 10.1007/978-94-017-9050-5_4. Subcell Biochem. 2014. PMID: 24962881 Review.
-
Receptor-selective retinoid agonists and teratogenic activity.Drug Metab Rev. 1996 Feb-May;28(1-2):105-19. doi: 10.3109/03602539608993994. Drug Metab Rev. 1996. PMID: 8744592 Review.
Cited by
-
Modular evolution of PGC-1alpha in vertebrates.J Mol Evol. 2010 May;70(5):492-505. doi: 10.1007/s00239-010-9347-x. Epub 2010 May 5. J Mol Evol. 2010. PMID: 20443112
-
Unveiling the role of RARs in stomach adenocarcinoma: clinical implications and prognostic biomarkers.Transl Cancer Res. 2024 Aug 31;13(8):3974-3995. doi: 10.21037/tcr-23-2154. Epub 2024 Aug 12. Transl Cancer Res. 2024. PMID: 39262490 Free PMC article.
-
Simulated evolution of protein-protein interaction networks with realistic topology.PLoS One. 2012;7(6):e39052. doi: 10.1371/journal.pone.0039052. Epub 2012 Jun 29. PLoS One. 2012. PMID: 22768057 Free PMC article.
-
The phytoestrogen genistein affects zebrafish development through two different pathways.PLoS One. 2009;4(3):e4935. doi: 10.1371/journal.pone.0004935. Epub 2009 Mar 25. PLoS One. 2009. PMID: 19319186 Free PMC article.
-
Lineage-specific duplication of amphioxus retinoic acid degrading enzymes (CYP26) resulted in sub-functionalization of patterning and homeostatic roles.BMC Evol Biol. 2017 Jan 19;17(1):24. doi: 10.1186/s12862-016-0863-1. BMC Evol Biol. 2017. PMID: 28103795 Free PMC article.
References
-
- Ohno S. Evolution by gene duplication. Heidelberg: Springer-Verlag; 1970.
-
- Holland PW, Garcia-Fernandez J, Williams NA, Sidow A. Gene duplications and the origins of vertebrate development. Dev Suppl. 1994;1994:125–133. - PubMed
-
- Taylor JS, Raes J. Duplication and divergence: The evolution of new genes and old ideas. Annu Rev Genet. 2004;38:615–643. - PubMed
-
- Dehal P, Boore JL. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 2005;3:e314. DOI: 10.1371/journal.pbio.0030314. - DOI - PMC - PubMed
-
- Delsuc F, Brinkmann H, Chourrout D, Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439:965–968. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

