DNA microarray technologies for measuring protein-DNA interactions
- PMID: 16839757
- PMCID: PMC2727741
- DOI: 10.1016/j.copbio.2006.06.015
DNA microarray technologies for measuring protein-DNA interactions
Abstract
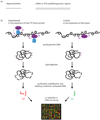
DNA-binding proteins have key roles in many cellular processes, including transcriptional regulation and replication. Microarray-based technologies permit the high-throughput identification of binding sites and enable the functional roles of these binding proteins to be elucidated. In particular, microarray readout either of chromatin immunoprecipitated DNA-bound proteins (ChIP-chip) or of DNA adenine methyltransferase fusion proteins (DamID) enables the identification of in vivo genomic target sites of proteins. A complementary approach to analyse the in vitro binding of proteins directly to double-stranded DNA microarrays (protein binding microarrays; PBMs), permits rapid characterization of their DNA binding site sequence specificities. Recent advances in DNA microarray synthesis technologies have facilitated the definition of DNA-binding sites at much higher resolution and coverage, and advances in these and emerging technologies will further increase the efficiencies of these exciting new approaches.
Figures


Similar articles
-
Protein binding microarrays for the characterization of DNA-protein interactions.Adv Biochem Eng Biotechnol. 2007;104:65-85. doi: 10.1007/10_025. Adv Biochem Eng Biotechnol. 2007. PMID: 17290819 Free PMC article. Review.
-
ChIP-chip to analyze the binding of replication proteins to chromatin using oligonucleotide DNA microarrays.Methods Mol Biol. 2009;521:255-78. doi: 10.1007/978-1-60327-815-7_14. Methods Mol Biol. 2009. PMID: 19563111
-
Protein binding microarrays (PBMs) for rapid, high-throughput characterization of the sequence specificities of DNA binding proteins.Methods Mol Biol. 2006;338:245-60. doi: 10.1385/1-59745-097-9:245. Methods Mol Biol. 2006. PMID: 16888363 Free PMC article.
-
DamID: mapping of in vivo protein-genome interactions using tethered DNA adenine methyltransferase.Methods Enzymol. 2006;410:342-59. doi: 10.1016/S0076-6879(06)10016-6. Methods Enzymol. 2006. PMID: 16938559 Review.
-
Analysis of Protein-DNA Interaction by Chromatin Immunoprecipitation and DNA Tiling Microarray (ChIP-on-chip).Methods Mol Biol. 2018;1689:43-51. doi: 10.1007/978-1-4939-7380-4_4. Methods Mol Biol. 2018. PMID: 29027163
Cited by
-
Empirical methods for controlling false positives and estimating confidence in ChIP-Seq peaks.BMC Bioinformatics. 2008 Dec 5;9:523. doi: 10.1186/1471-2105-9-523. BMC Bioinformatics. 2008. PMID: 19061503 Free PMC article.
-
UniPROBE: an online database of protein binding microarray data on protein-DNA interactions.Nucleic Acids Res. 2009 Jan;37(Database issue):D77-82. doi: 10.1093/nar/gkn660. Epub 2008 Oct 8. Nucleic Acids Res. 2009. PMID: 18842628 Free PMC article.
-
Single-target regulators form a minor group of transcription factors in Escherichia coli K-12.Nucleic Acids Res. 2018 May 4;46(8):3921-3936. doi: 10.1093/nar/gky138. Nucleic Acids Res. 2018. PMID: 29529243 Free PMC article.
-
Exact p-value calculation for heterotypic clusters of regulatory motifs and its application in computational annotation of cis-regulatory modules.Algorithms Mol Biol. 2007 Oct 10;2:13. doi: 10.1186/1748-7188-2-13. Algorithms Mol Biol. 2007. PMID: 17927813 Free PMC article.
-
De novo ChIP-seq analysis.Genome Biol. 2015 Sep 23;16(1):205. doi: 10.1186/s13059-015-0756-4. Genome Biol. 2015. PMID: 26400819 Free PMC article.
References
-
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. - PubMed
-
- Lieb JD, Liu X, Botstein D, Brown PO. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat Genet. 2001;28:327–334. - PubMed
-
- Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. - PubMed
-
- Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

