Promoter activation by the varicella-zoster virus major transactivator IE62 and the cellular transcription factor USF
- PMID: 16840315
- PMCID: PMC1563731
- DOI: 10.1128/JVI.00309-06
Promoter activation by the varicella-zoster virus major transactivator IE62 and the cellular transcription factor USF
Abstract
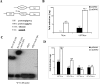
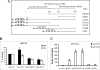
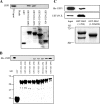
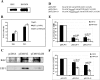
The varicella-zoster virus major transactivator, IE62, can activate expression from homologous and heterologous promoters. High levels of IE62-mediated activation appear to involve synergy with cellular transcription factors. The work presented here focuses on functional interactions of IE62 with the ubiquitously expressed cellular factor USF. We have found that USF can synergize with IE62 to a similar extent on model minimal promoters and the complex native ORF28/29 regulatory element, neither of which contains a consensus IE62 binding site. Using Gal4 fusion constructs, we have found that the activation domain of USF1 is necessary and sufficient for synergistic activation with IE62. We have mapped the regions of USF and IE62 required for direct physical interaction. Deletion of the required region within IE62 does not ablate synergistic activation but does influence its efficiency depending on promoter architecture. Both proteins stabilize/increase binding of TATA binding protein/TFIID to promoter elements. These findings suggest a novel mechanism for the observed synergistic activation which requires neither site-specific IE62 binding to the promoter nor a direct physical interaction with USF.
Figures



Similar articles
-
Cellular factors and IE62 activation of VZV promoters.J Med Virol. 2003;70 Suppl 1:S90-4. doi: 10.1002/jmv.10328. J Med Virol. 2003. PMID: 12627495
-
The DNA element controlling expression of the varicella-zoster virus open reading frame 28 and 29 genes consists of two divergent unidirectional promoters which have a common USF site.J Virol. 2004 Oct;78(20):10939-52. doi: 10.1128/JVI.78.20.10939-10952.2004. J Virol. 2004. PMID: 15452214 Free PMC article.
-
The TATA motif specifies the differential activation of minimal promoters by varicella zoster virus immediate-early regulatory protein IE62.J Biol Chem. 2000 Jan 7;275(1):487-96. doi: 10.1074/jbc.275.1.487. J Biol Chem. 2000. PMID: 10617643
-
Varicella-zoster virus IE62 protein utilizes the human mediator complex in promoter activation.J Virol. 2008 Dec;82(24):12154-63. doi: 10.1128/JVI.01693-08. Epub 2008 Oct 8. J Virol. 2008. PMID: 18842726 Free PMC article.
-
Roles of cellular transcription factors in VZV replication.Curr Top Microbiol Immunol. 2010;342:43-65. doi: 10.1007/82_2010_42. Curr Top Microbiol Immunol. 2010. PMID: 20490778 Review.
Cited by
-
The ubiquitous cellular transcriptional factor USF targets the varicella-zoster virus open reading frame 10 promoter and determines virulence in human skin xenografts in SCIDhu mice in vivo.J Virol. 2007 Apr;81(7):3229-39. doi: 10.1128/JVI.02537-06. Epub 2007 Jan 24. J Virol. 2007. PMID: 17251302 Free PMC article.
-
Role of the IE62 consensus binding site in transactivation by the varicella-zoster virus IE62 protein.J Virol. 2010 Apr;84(8):3767-79. doi: 10.1128/JVI.02522-09. Epub 2010 Feb 3. J Virol. 2010. PMID: 20130051 Free PMC article.
-
Mutational analysis of varicella-zoster virus (VZV) immediate early protein (IE62) subdomains and their importance in viral replication.Virology. 2016 May;492:82-91. doi: 10.1016/j.virol.2016.02.012. Epub 2016 Feb 23. Virology. 2016. PMID: 26914506 Free PMC article.
-
Cellular transcription factors Sp1 and Sp3 suppress varicella-zoster virus origin-dependent DNA replication.J Virol. 2008 Dec;82(23):11723-33. doi: 10.1128/JVI.01322-08. Epub 2008 Sep 24. J Virol. 2008. PMID: 18815296 Free PMC article.
-
Identification and functional characterization of the Varicella zoster virus ORF11 gene product.Virology. 2011 Mar 30;412(1):156-66. doi: 10.1016/j.virol.2010.12.055. Epub 2011 Jan 26. Virology. 2011. PMID: 21276599 Free PMC article.
References
-
- Argaw, T., J. I. Cohen, M. Klutch, K. Lekstrom, T. Yoshikawa, Y. Asano, and P. R. Krause. 2000. Nucleotide sequences that distinguish Oka vaccine from parental Oka and other varicella-zoster virus isolates. J. Infect. Dis. 181:1153-1157. - PubMed
-
- Betz, J. L., and S. G. Wydoski. 1993. Functional interaction of varicella-zoster virus gene 62 protein with the DNA sequence bound by herpes simplex virus ICP4 protein. Virology 195:793-797. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

