The NS5A protein of bovine viral diarrhea virus contains an essential zinc-binding site similar to that of the hepatitis C virus NS5A protein
- PMID: 16840325
- PMCID: PMC1563740
- DOI: 10.1128/JVI.00358-06
The NS5A protein of bovine viral diarrhea virus contains an essential zinc-binding site similar to that of the hepatitis C virus NS5A protein
Abstract
The recent demonstration that the NS5A protein of hepatitis C virus (HCV) contains an unconventional zinc-binding site with the format Cx(17)CxCx(20)C and the presence of a similar sequence element in the NS5A proteins of members of the Pestivirus genus has led to the hypothesis that the NS5A protein of the pestivirus bovine viral diarrhea virus (BVDV) is a zinc-binding protein. A method for the expression and partial purification of BVDV NS5A was developed, and the partially purified protein was analyzed for zinc content by atomic absorption spectroscopy. BVDV NS5A was found to coordinate a single zinc atom per protein molecule. Mutation of any of the four cysteines of the predicted zinc-binding motif eliminated zinc coordination. Furthermore, analysis of mutations at these cysteine residues in the context of a BVDV replicon system indicated that these residues were absolutely essential for RNA replication. The recently determined crystal structure of the N-terminal zinc-binding domain of the HCV NS5A protein, combined with secondary structure predictions of the region surrounding the mapped BVDV zinc-binding region, indicates that the BVDV zinc-binding motif fits the general template Cx(22)CxCx(24)C and likely comprises a three-stranded antiparallel beta-sheet fold. These data highlight the similarities between the Hepacivirus and Pestivirus NS5A proteins and suggest that both proteins perform a not-yet-defined function in RNA replication that requires coordination of a single zinc atom.
Figures

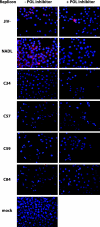
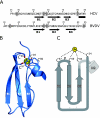
Similar articles
-
The NS5A protein of hepatitis C virus is a zinc metalloprotein.J Biol Chem. 2004 Nov 19;279(47):48576-87. doi: 10.1074/jbc.M407787200. Epub 2004 Aug 31. J Biol Chem. 2004. PMID: 15339921
-
Temporal modulation of an autoprotease is crucial for replication and pathogenicity of an RNA virus.J Virol. 2004 Oct;78(19):10765-75. doi: 10.1128/JVI.78.19.10765-10775.2004. J Virol. 2004. PMID: 15367643 Free PMC article.
-
Structure of the zinc-binding domain of an essential component of the hepatitis C virus replicase.Nature. 2005 May 19;435(7040):374-9. doi: 10.1038/nature03580. Nature. 2005. PMID: 15902263 Free PMC article.
-
Non-structural proteins of bovine viral diarrhea virus.Virus Genes. 2022 Dec;58(6):491-500. doi: 10.1007/s11262-022-01914-8. Epub 2022 May 25. Virus Genes. 2022. PMID: 35614328 Free PMC article. Review.
-
Hepatitis C virus NS5A: tales of a promiscuous protein.J Gen Virol. 2004 Sep;85(Pt 9):2485-2502. doi: 10.1099/vir.0.80204-0. J Gen Virol. 2004. PMID: 15302943 Review.
Cited by
-
Augmentation of 3β-hydroxysteroid-Δ24 Reductase (DHCR24) Expression Induced by Bovine Viral Diarrhea Virus Infection Facilitates Viral Replication via Promoting Cholesterol Synthesis.J Virol. 2022 Dec 21;96(24):e0149222. doi: 10.1128/jvi.01492-22. Epub 2022 Dec 5. J Virol. 2022. PMID: 36468862 Free PMC article.
-
Rab18 binds to classical swine fever virus NS5A and mediates viral replication and assembly in swine umbilical vein endothelial cells.Virulence. 2020 Dec;11(1):489-501. doi: 10.1080/21505594.2020.1767356. Virulence. 2020. PMID: 32419589 Free PMC article.
-
Stability of the resistance to the thiosemicarbazone derived from 5,6-dimethoxy-1-indanone, a non-nucleoside polymerase inhibitor of bovine viral diarrhea virus.PLoS One. 2014 Jun 20;9(6):e100528. doi: 10.1371/journal.pone.0100528. eCollection 2014. PLoS One. 2014. PMID: 24950191 Free PMC article.
-
Classical swine fever virus NS5A protein localizes to endoplasmic reticulum and induces oxidative stress in vascular endothelial cells.Virus Genes. 2012 Oct;45(2):274-82. doi: 10.1007/s11262-012-0773-2. Epub 2012 Jun 21. Virus Genes. 2012. PMID: 22718084
-
The unexpected roles of eukaryotic translation elongation factors in RNA virus replication and pathogenesis.Microbiol Mol Biol Rev. 2013 Jun;77(2):253-66. doi: 10.1128/MMBR.00059-12. Microbiol Mol Biol Rev. 2013. PMID: 23699257 Free PMC article. Review.
References
-
- Bazan, J. F., and R. J. Fletterick. 1989. Detection of a trypsin-like serine protease domain in flaviviruses and pestiviruses. Virology 171:637-639. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

