Characterization of the archaeal thermophile Sulfolobus turreted icosahedral virus validates an evolutionary link among double-stranded DNA viruses from all domains of life
- PMID: 16840341
- PMCID: PMC1563717
- DOI: 10.1128/JVI.00522-06
Characterization of the archaeal thermophile Sulfolobus turreted icosahedral virus validates an evolutionary link among double-stranded DNA viruses from all domains of life
Abstract
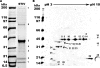
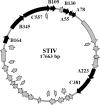
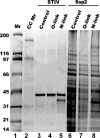
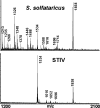
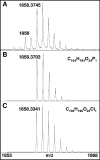
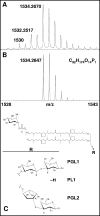
Icosahedral nontailed double-stranded DNA (dsDNA) viruses are present in all three domains of life, leading to speculation about a common viral ancestor that predates the divergence of Eukarya, Bacteria, and Archaea. This suggestion is supported by the shared general architecture of this group of viruses and the common fold of their major capsid protein. However, limited information on the diversity and replication of archaeal viruses, in general, has hampered further analysis. Sulfolobus turreted icosahedral virus (STIV), isolated from a hot spring in Yellowstone National Park, was the first icosahedral virus with an archaeal host to be described. Here we present a detailed characterization of the components forming this unusual virus. Using a proteomics-based approach, we identified nine viral and two host proteins from purified STIV particles. Interestingly, one of the viral proteins originates from a reading frame lacking a consensus start site. The major capsid protein (B345) was found to be glycosylated, implying a strong similarity to proteins from other dsDNA viruses. Sequence analysis and structural predication of virion-associated viral proteins suggest that they may have roles in DNA packaging, penton formation, and protein-protein interaction. The presence of an internal lipid layer containing acidic tetraether lipids has also been confirmed. The previously presented structural models in conjunction with the protein, lipid, and carbohydrate information reported here reveal that STIV is strikingly similar to viruses associated with the Bacteria and Eukarya domains of life, further strengthening the hypothesis for a common ancestor of this group of dsDNA viruses from all domains of life.
Figures
Similar articles
-
Structure of A197 from Sulfolobus turreted icosahedral virus: a crenarchaeal viral glycosyltransferase exhibiting the GT-A fold.J Virol. 2006 Aug;80(15):7636-44. doi: 10.1128/JVI.00567-06. J Virol. 2006. PMID: 16840342 Free PMC article.
-
Structure-Based Mutagenesis of Sulfolobus Turreted Icosahedral Virus B204 Reveals Essential Residues in the Virion-Associated DNA-Packaging ATPase.J Virol. 2015 Dec 23;90(6):2729-39. doi: 10.1128/JVI.02435-15. J Virol. 2015. PMID: 26699645 Free PMC article.
-
Development of a genetic system for the archaeal virus Sulfolobus turreted icosahedral virus (STIV).Virology. 2011 Jun 20;415(1):6-11. doi: 10.1016/j.virol.2011.03.023. Epub 2011 Apr 15. Virology. 2011. PMID: 21496857
-
Structure and cell biology of archaeal virus STIV.Curr Opin Virol. 2012 Apr;2(2):122-7. doi: 10.1016/j.coviro.2012.01.007. Epub 2012 Mar 20. Curr Opin Virol. 2012. PMID: 22482708 Free PMC article. Review.
-
Genetics, biochemistry and structure of the archaeal virus STIV.Biochem Soc Trans. 2009 Feb;37(Pt 1):114-7. doi: 10.1042/BST0370114. Biochem Soc Trans. 2009. PMID: 19143613 Review.
Cited by
-
The Prevalence of STIV c92-Like Proteins in Acidic Thermal Environments.Adv Virol. 2011;2011:650930. doi: 10.1155/2011/650930. Epub 2011 Jul 14. Adv Virol. 2011. PMID: 22312348 Free PMC article.
-
Viruses of the Turriviridae: an emerging model system for studying archaeal virus-host interactions.Front Microbiol. 2023 Sep 21;14:1258997. doi: 10.3389/fmicb.2023.1258997. eCollection 2023. Front Microbiol. 2023. PMID: 37808280 Free PMC article. Review.
-
HCIV-1 and Other Tailless Icosahedral Internal Membrane-Containing Viruses of the Family Sphaerolipoviridae.Viruses. 2017 Feb 18;9(2):32. doi: 10.3390/v9020032. Viruses. 2017. PMID: 28218714 Free PMC article. Review.
-
Functional interplay between a virus and the ESCRT machinery in archaea.Proc Natl Acad Sci U S A. 2013 Jun 25;110(26):10783-7. doi: 10.1073/pnas.1301605110. Epub 2013 Jun 10. Proc Natl Acad Sci U S A. 2013. PMID: 23754419 Free PMC article.
-
Structure of A197 from Sulfolobus turreted icosahedral virus: a crenarchaeal viral glycosyltransferase exhibiting the GT-A fold.J Virol. 2006 Aug;80(15):7636-44. doi: 10.1128/JVI.00567-06. J Virol. 2006. PMID: 16840342 Free PMC article.
References
-
- Abrescia, N. G., J. J. Cockburn, J. M. Grimes, G. C. Sutton, J. M. Diprose, S. J. Butcher, S. D. Fuller, C. San Martin, R. M. Burnett, D. I. Stuart, D. H. Bamford, and J. K. Bamford. 2004. Insights into assembly from structural analysis of bacteriophage PRD1. Nature 432:68-74. - PubMed
-
- Bamford, D. H., J. M. Grimes, and D. I. Staurt. 2005. What does structure tell us about virus evolution? Curr. Opin. Struct. Biol. 15:655-663. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

