Effective suppression of human immunodeficiency virus type 1 through a combination of short- or long-hairpin RNAs targeting essential sequences for retroviral integration
- PMID: 16840344
- PMCID: PMC1563699
- DOI: 10.1128/JVI.00078-06
Effective suppression of human immunodeficiency virus type 1 through a combination of short- or long-hairpin RNAs targeting essential sequences for retroviral integration
Abstract
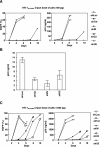
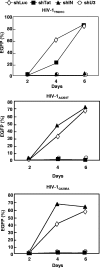
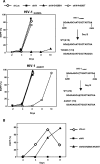
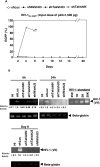
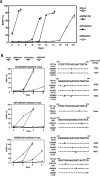
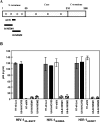
Small interfering RNA (siRNA) could provide a new therapeutic approach to treating human immunodeficiency virus type 1 (HIV-1) infection. For long-term suppression of HIV-1, emergence of siRNA escape variants must be controlled. Here, we constructed lentiviral vectors encoding short-hairpin RNAs (shRNA) corresponding to conserved target sequences within the integrase (int) and the attachment site (att) genes, both of which are essential for HIV-1 integration. Compared to shRNA targeting of the HIV-1 transcription factor tat (shTat), shRNA against int (shIN) or the U3 region of att (shU3) showed a more potent inhibitory effect on HIV-1 replication in human CD4+ T cells. Infection with a high dose of HIV-1 resulted in the emergence of escape mutants during long-term culture. Of note, limited genetic variation was observed in the viruses resistant to shIN. A combination of shINs against wild-type and escape mutant sequences had a negative effect on their antiviral activities, indicating a potentially detrimental effect when administering multiple shRNA targeting the same region to combat HIV-1 variants. The combination of shIN and shU3 att exhibited the strongest anti-HIV-1 activity, as seen by complete abrogation of viral DNA synthesis and viral integration. In addition, a modified long-hairpin RNA spanning the 50 nucleotides in the shIN target region effectively suppressed wild-type and shIN-resistant mutant HIV-1. These results suggest that targeting of incoming viral RNA before proviral DNA formation occurs through the use of nonoverlapping multiple siRNAs is a potent approach to achieving sustained, efficient suppression of highly mutable viruses, such as HIV-1.
Figures


References
-
- Banerjea, A., M. J. Li, G. Bauer, L. Remling, N. S. Lee, J. Rossi, and R. Akkina. 2003. Inhibition of HIV-1 by lentiviral vector-transduced siRNAs in T lymphocytes differentiated in SCID-hu mice and CD34+ progenitor cell-derived macrophages. Mol. Ther. 8:62-71. - PubMed
-
- Bennasser, Y., S. Y. Le, M. Benkirane, and K. T. Jeang. 2005. Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity 22:607-619. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

