Locus-specific gene expression pattern suggests a unique propagation strategy for a giant algal virus
- PMID: 16840348
- PMCID: PMC1563701
- DOI: 10.1128/JVI.00491-06
Locus-specific gene expression pattern suggests a unique propagation strategy for a giant algal virus
Abstract
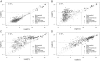
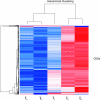
Emiliania huxleyi virus strain 86 is the largest algal virus sequenced to date and is unique among the Phycodnaviridae since its genome is predicted to contain six RNA polymerase subunit genes. We have used a virus microarray to profile the temporal transcription strategy of this unusual virus during infection. There are two distinct transcription phases to the infection process. The primary phase is dominated by a group of coding sequences (CDSs) expressed by 1 h postinfection that are localized to a subregion of the genome. The CDS of the primary group have no database homologues, and each is associated with a unique promoter element. The remainder of the CDSs are expressed in a secondary phase between 2 and 4 hours postinfection. Compartmentalized transcription of the two distinctive phases is discussed. We hypothesize that immediately after infection the nucleic acid of the virus targets the host nucleus, where primary-phase genes are transcribed by host RNA polymerase which recognizes the viral promoter. Secondary-phase transcription may then be conducted in the cytoplasm.
Figures


References
-
- Allen, M. J., D. C. Schroeder, M. T. Holden, and W. H. Wilson. 2006. Evolutionary history of the coccolithoviridae. Mol. Biol. Evol. 23:86-92. - PubMed
-
- Allen, M. J., D. C. Schroeder, and W. H. Wilson. 2006. Preliminary characterisation of repeat families in the genome of EhV-86, a giant algal virus that infects the marine microalga Emiliania huxleyi. Arch. Virol. 151:525-535. - PubMed
-
- Amici, C., A. Rossi, A. Costanzo, S. Ciafre, B. Marinari, M. Balsamo, M. Levrero, and M. G. Santoro. 2006. Herpes simplex virus disrupts NF-κB regulation by blocking its recruitment on the IκBα promoter and directing the factor on viral genes. J. Biol. Chem. 281:7110-7117. - PubMed
-
- Chambers, J., A. Angulo, D. Amaratunga, H. Q. Guo, Y. Jiang, J. S. Wan, A. Bittner, K. Frueh, M. R. Jackson, P. A. Peterson, M. G. Erlander, and P. Ghazal. 1999. DNA microarrays of the complex human cytomegalovirus genome: profiling kinetic class with drug sensitivity of viral gene expression. J. Virol. 73:5757-5766. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

