A poxvirus host range protein, CP77, binds to a cellular protein, HMG20A, and regulates its dissociation from the vaccinia virus genome in CHO-K1 cells
- PMID: 16840350
- PMCID: PMC1563734
- DOI: 10.1128/JVI.00207-06
A poxvirus host range protein, CP77, binds to a cellular protein, HMG20A, and regulates its dissociation from the vaccinia virus genome in CHO-K1 cells
Abstract
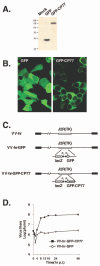
Vaccinia virus does not grow in Chinese hamster ovary (CHO-K1) cells in the absence of a viral host range factor, cowpox protein CP77. In this study, CP77 was fused to the C terminus of green fluorescence protein (GFP-CP77) and a series of nested deletion mutants of GFP-CP77 was constructed for insertion into a vaccinia virus host range mutant, VV-hr, and expressed from a viral early promoter. Deletion mapping analyses demonstrated that the N-terminal 352 amino acids of CP77 were sufficient to support vaccinia virus growth in CHO-K1 cells, whereas the C-terminal residues 353 to 668 were dispensable. In yeast two-hybrid analyses, CP77 bound to a cellular protein, HMG20A, and GST pulldown analyses showed that residues 1 to 234 of CP77 were sufficient for this interaction. After VV-hr virus infection of CHO-K1 cells, HMG20A was translocated from the nucleus to viral factories and bound to the viral genome via the HMG box region. In control VV-hr-infected CHO-K1 cells, binding of HMG20A to the viral genome persisted from 2 to 8 h postinfection (h p.i.); in contrast, when CP77 was expressed, the association of HMG20A with viral genome was transient, with little HMG20A remaining bound at 8 h p.i. This indicates that dissociation of HMG20A from viral factories correlates well with CP77 host range activity in CHO-K1 cells. Finally, in cells expressing a CP77 deletion protein (amino acids 277 to 668) or a DeltaANK5 mutant that did not support vaccinia virus growth and did not contain the HMG20A binding site, HMG20A remained bound to viral DNA, demonstrating that the binding of CP77 to HMG20A is essential for its host range function. In summary, our data revealed that a novel cellular protein, HMG20A, the dissociation of which from viral DNA is regulated by CP77, providing the first cellular target regulated by viral host range CP77 protein.
Figures









Similar articles
-
The cowpox virus host range gene, CP77, affects phosphorylation of eIF2 alpha and vaccinia viral translation in apoptotic HeLa cells.Virology. 2004 Nov 10;329(1):199-212. doi: 10.1016/j.virol.2004.07.032. Virology. 2004. PMID: 15476887
-
Generation and characterization of a Cowpox virus mutant lacking host range factor CP77.Virus Res. 2012 Sep;168(1-2):23-32. doi: 10.1016/j.virusres.2012.06.005. Epub 2012 Jun 13. Virus Res. 2012. PMID: 22705200
-
Complementation of a vaccinia virus host-range K1L gene deletion by the nonhomologous CP77 gene.Virology. 1996 Aug 1;222(1):75-86. doi: 10.1006/viro.1996.0399. Virology. 1996. PMID: 8806489
-
[Research progress in the structure and fuction of Orthopoxvirus host range genes].Bing Du Xue Bao. 2013 Jun;29(4):437-41. Bing Du Xue Bao. 2013. PMID: 23895011 Review. Chinese.
-
Poxvirus proteomics and virus-host protein interactions.Microbiol Mol Biol Rev. 2009 Dec;73(4):730-49. doi: 10.1128/MMBR.00026-09. Microbiol Mol Biol Rev. 2009. PMID: 19946139 Free PMC article. Review.
Cited by
-
Evaluation of Viral Genome Copies Within Viral Factories on Different DNA Viruses.J Histochem Cytochem. 2018 May;66(5):359-365. doi: 10.1369/0022155417749490. Epub 2018 Jan 3. J Histochem Cytochem. 2018. PMID: 29298122 Free PMC article.
-
Vaccinia virus DNA ligase recruits cellular topoisomerase II to sites of viral replication and assembly.J Virol. 2008 Jun;82(12):5922-32. doi: 10.1128/JVI.02723-07. Epub 2008 Apr 16. J Virol. 2008. PMID: 18417590 Free PMC article.
-
Identification of CP77 as the Third Orthopoxvirus SAMD9 and SAMD9L Inhibitor with Unique Specificity for a Rodent SAMD9L.J Virol. 2019 May 29;93(12):e00225-19. doi: 10.1128/JVI.00225-19. Print 2019 Jun 15. J Virol. 2019. PMID: 30918078 Free PMC article.
-
Genomic analysis reveals an exogenous viral symbiont with dual functionality in parasitoid wasps and their hosts.PLoS Pathog. 2020 Nov 30;16(11):e1009069. doi: 10.1371/journal.ppat.1009069. eCollection 2020 Nov. PLoS Pathog. 2020. PMID: 33253317 Free PMC article.
-
The interaction with HMG20a/b proteins suggests a potential role for beta-dystrobrevin in neuronal differentiation.J Biol Chem. 2010 Aug 6;285(32):24740-50. doi: 10.1074/jbc.M109.090654. Epub 2010 Jun 8. J Biol Chem. 2010. PMID: 20530487 Free PMC article.
References
-
- Ali, A. N., P. C. Turner, M. A. Brooks, and R. W. Moyer. 1994. The SP-1 gene of rabbitpox virus determines host range and is required for hemorrhagic pock formation. Virology 202:305-314. - PubMed
-
- Baroudy, B. M., S. Venkatesan, and B. Moss. 1983. Structure and replication of vaccinia virus telomeres. Cold Spring Harbor Symp. Quant. Biol. 47:723-729. - PubMed
-
- Barry, M., S. Hnatiuk, K. Mossman, S. F. Lee, L. Boshkov, and G. McFadden. 1997. The myxoma virus M-T4 gene encodes a novel RDEL-containing protein that is retained within the endoplasmic reticulum and is important for the productive infection of lymphocytes. Virology 239:360-377. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

