Genomic DNA functions as a universal external standard in quantitative real-time PCR
- PMID: 16840529
- PMCID: PMC1524913
- DOI: 10.1093/nar/gkl400
Genomic DNA functions as a universal external standard in quantitative real-time PCR
Erratum in
- Nucleic Acids Res. 2006;34(22):6718
Abstract
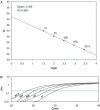
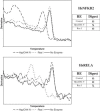
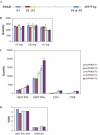
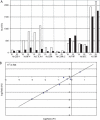
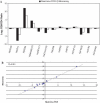
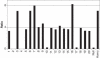
Real-time quantitative PCR (qPCR) is a powerful tool for quantifying specific DNA target sequences. Although determination of relative quantity is widely accepted as a reliable means of measuring differences between samples, there are advantages to being able to determine the absolute copy numbers of a given target. One approach to absolute quantification relies on construction of an accurate standard curve using appropriate external standards of known concentration. We have validated the use of tissue genomic DNA as a universal external standard to facilitate quantification of any target sequence contained in the genome of a given species, addressing several key technical issues regarding its use. This approach was applied to validate mRNA expression of gene candidates identified from microarray data and to determine gene copies in transgenic mice. A simple method that can assist achieving absolute quantification of gene expression would broadly enhance the uses of real-time qPCR and in particular, augment the evaluation of global gene expression studies.
Figures
References
-
- Rose'Meyer R.B., Mellick A.S., Garnham B.G., Harrison G.J., Massa H.M., Griffiths L.R. The measurement of adenosine and estrogen receptor expression in rat brains following ovariectomy using quantitative PCR analysis. Brain Res. Brain Res. Protoc. 2003;11:9–18. - PubMed
-
- Li X., Wang X. Application of real-time polymerase chain reaction for the quantitation of interleukin-1beta mRNA upregulation in brain ischemic tolerance. Brain Res. Brain Res. Protoc. 2000;5:211–217. - PubMed
-
- Yip S.P., Lee S.Y., To S.S., Wong M.L. Improved real-time PCR assay for homogeneous multiplex genotyping of four CYP2C9 alleles with hybridization probes. Clin. Chem. 2003;49:2109–2111. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

