Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly
- PMID: 16840558
- PMCID: PMC1544092
- DOI: 10.1073/pnas.0602818103
Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly
Abstract
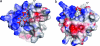
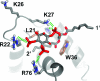
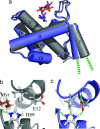
During the late phase of HIV type 1 (HIV-1) replication, newly synthesized retroviral Gag proteins are targeted to the plasma membrane of most hematopoietic cell types, where they colocalize at lipid rafts and assemble into immature virions. Membrane binding is mediated by the matrix (MA) domain of Gag, a 132-residue polypeptide containing an N-terminal myristyl group that can adopt sequestered and exposed conformations. Although exposure is known to promote membrane binding, the mechanism by which Gag is targeted to specific membranes has yet to be established. Recent studies have shown that phosphatidylinositol (PI) 4,5-bisphosphate [PI(4,5)P(2)], a factor that regulates localization of cellular proteins to the plasma membrane, also regulates Gag localization and assembly. Here we show that PI(4,5)P(2) binds directly to HIV-1 MA, inducing a conformational change that triggers myristate exposure. Related phosphatidylinositides PI, PI(3)P, PI(4)P, PI(5)P, and PI(3,5)P(2) do not bind MA with significant affinity or trigger myristate exposure. Structural studies reveal that PI(4,5)P(2) adopts an "extended lipid" conformation, in which the inositol head group and 2'-fatty acid chain bind to a hydrophobic cleft, and the 1'-fatty acid and exposed myristyl group bracket a conserved basic surface patch previously implicated in membrane binding. Our findings indicate that PI(4,5)P(2) acts as both a trigger of the myristyl switch and a membrane anchor and suggest a potential mechanism for targeting Gag to membrane rafts.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures

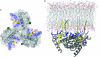
Comment in
-
HIV-1 Gag: flipped out for PI(4,5)P(2).Proc Natl Acad Sci U S A. 2006 Jul 25;103(30):11101-2. doi: 10.1073/pnas.0604715103. Epub 2006 Jul 17. Proc Natl Acad Sci U S A. 2006. PMID: 16847255 Free PMC article. No abstract available.
Similar articles
-
Point mutations in the HIV-1 matrix protein turn off the myristyl switch.J Mol Biol. 2007 Feb 16;366(2):574-85. doi: 10.1016/j.jmb.2006.11.068. Epub 2006 Dec 1. J Mol Biol. 2007. PMID: 17188710 Free PMC article.
-
Membrane Binding of HIV-1 Matrix Protein: Dependence on Bilayer Composition and Protein Lipidation.J Virol. 2016 Apr 14;90(9):4544-4555. doi: 10.1128/JVI.02820-15. Print 2016 May. J Virol. 2016. PMID: 26912608 Free PMC article.
-
Membrane binding and subcellular localization of retroviral Gag proteins are differentially regulated by MA interactions with phosphatidylinositol-(4,5)-bisphosphate and RNA.mBio. 2014 Dec 9;5(6):e02202. doi: 10.1128/mBio.02202-14. mBio. 2014. PMID: 25491356 Free PMC article.
-
[Membrane Binding of Retroviral Gag Proteins].Uirusu. 2014;64(2):155-64. doi: 10.2222/jsv.64.155. Uirusu. 2014. PMID: 26437838 Review. Japanese.
-
Intracellular trafficking of HIV-1 Gag: how Gag interacts with cell membranes and makes viral particles.AIDS Rev. 2005 Apr-Jun;7(2):84-91. AIDS Rev. 2005. PMID: 16092502 Review.
Cited by
-
Evidence in support of RNA-mediated inhibition of phosphatidylserine-dependent HIV-1 Gag membrane binding in cells.J Virol. 2013 Jun;87(12):7155-9. doi: 10.1128/JVI.00075-13. Epub 2013 Apr 3. J Virol. 2013. PMID: 23552424 Free PMC article.
-
Evaluation of phosphatidylinositol-4-kinase IIIα as a hepatitis C virus drug target.J Virol. 2012 Nov;86(21):11595-607. doi: 10.1128/JVI.01320-12. Epub 2012 Aug 15. J Virol. 2012. PMID: 22896614 Free PMC article.
-
NMR Studies of Retroviral Genome Packaging.Viruses. 2020 Sep 30;12(10):1115. doi: 10.3390/v12101115. Viruses. 2020. PMID: 33008123 Free PMC article. Review.
-
Effects of Membrane Charge and Order on Membrane Binding of the Retroviral Structural Protein Gag.J Virol. 2016 Sep 29;90(20):9518-32. doi: 10.1128/JVI.01102-16. Print 2016 Oct 15. J Virol. 2016. PMID: 27512076 Free PMC article.
-
Dynamic Association between HIV-1 Gag and Membrane Domains.Mol Biol Int. 2012;2012:979765. doi: 10.1155/2012/979765. Epub 2012 Jul 5. Mol Biol Int. 2012. PMID: 22830021 Free PMC article.
References
-
- Gheysen D., Jacobs E., de Foresta F., Thiriart C., Francotte M., Thines D., De Wilde M. Cell. 1989;59:103–112. - PubMed
-
- Wills J., Craven R. AIDS. 1991;5:639–654. - PubMed
-
- Freed E. O. Virology. 1998;251:1–15. - PubMed
-
- Briggs J. A. G., Simon M. N., Gross I., Krausslich H.-G., Fuller S. D., Vogt V. M., Johnson M. C. Nat. Struct. Mol. Biol. 2004;11:672–675. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

