Development and application of a suite of polysaccharide-degrading enzymes for analyzing plant cell walls
- PMID: 16844780
- PMCID: PMC1544100
- DOI: 10.1073/pnas.0604632103
Development and application of a suite of polysaccharide-degrading enzymes for analyzing plant cell walls
Abstract
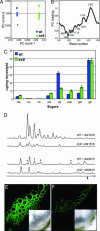
To facilitate analysis of plant cell wall polysaccharide structure and composition, we cloned 74 genes encoding polysaccharide-degrading enzymes from Aspergillus nidulans, Aspergillus fumigatus, and Neurospora crassa and expressed the genes as secreted proteins with C-terminal Myc and 6x His tags. Most of the recombinant enzymes were active in enzyme assays, and optima for pH and temperature were established. A subset of the enzymes was used to fragment polysaccharides from the irregular xylem 9 (irx9) mutant of Arabidopsis. The analysis revealed a decrease in the abundance of xylan in the mutant, indicating that the IRX9 gene, which encodes a putative family 43 glycosyltransferase, is required for xylan synthesis.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures
References
-
- Coimbra M. A., Delgadillo I., Waldron K. W., Selvendran R. R. In: Modern Methods of Plant Analysis: Plant Cell Wall Analysis. Linskens H. F., Jackson J. F, editors. Vol. 17. Berlin: Springer; 1996. pp. 19–44.
-
- Strasser G. R., Amado R. Carbohydr. Polymers. 2002;48:263–269.
-
- Deery M. J, Stimson E, Chappell C. G. Rapid Commun. Mass Spectrom. 2001;15:2273–2283. - PubMed
-
- Cohen A., Schagerlof H., Nilsson C., Melander C., Tjerneld F., Gorton L. J. Chromatogr. A. 2004;1029:87–95. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

