Regulatory protein that inhibits both synthesis and use of the target protein controls flagellar phase variation in Salmonella enterica
- PMID: 16844786
- PMCID: PMC1544088
- DOI: 10.1073/pnas.0602127103
Regulatory protein that inhibits both synthesis and use of the target protein controls flagellar phase variation in Salmonella enterica
Abstract
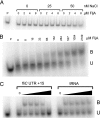
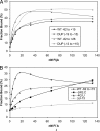
Flagellin is a major surface antigen for many bacterial species. The pathogen Salmonella enterica switches between two alternative, antigenic forms of its flagellin filament protein, either type B or C. This switching (flagellar phase variation) is achieved by stochastic inversion of a promoter that produces both type B flagellin (FljB) and an inhibitor (FljA) of type C flagellin formation. When the fljB-fljA operon is expressed, only type B flagella are produced; when the operon is not transcribed, the gene for type C flagellin (fliC) is released from inhibition and forms type C flagella. Long thought to be a transcription repressor, the FljA inhibitor is shown here to block both translation and use of the FliC protein by binding to an mRNA region upstream from the translation start codon. Bypass mutants resistant to this inhibition alter this mRNA region, and some prevent FljA-RNA binding. Other bypass mutations are duplications within the leader mRNA that make FljA essential for FliC assembly. Certain bypass mutations allow FljA to block FliC-dependent motility without blocking production of the FliC protein, per se. Other mutations in the FliC mRNA leader block expression of the unlinked fljB gene. Results suggest that mRNAs for types B and C flagellin compete for occupancy of a site that directs the product toward assembly and that FljA influences this competition. This mechanism may serve to prevent assembly of flagella with a mixture of subunit types, especially during periods of switching from one type to the other.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

