The 2.7-Angstrom crystal structure of a 194-kDa homodimeric fragment of the 6-deoxyerythronolide B synthase
- PMID: 16844787
- PMCID: PMC1636687
- DOI: 10.1073/pnas.0601924103
The 2.7-Angstrom crystal structure of a 194-kDa homodimeric fragment of the 6-deoxyerythronolide B synthase
Abstract
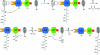
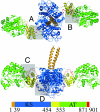
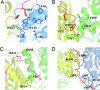
The x-ray crystal structure of a 194-kDa fragment from module 5 of the 6-deoxyerythronolide B synthase has been solved at 2.7 Angstrom resolution. Each subunit of the homodimeric protein contains a full-length ketosynthase (KS) and acyl transferase (AT) domain as well as three flanking "linkers." The linkers are structurally well defined and contribute extensively to intersubunit or interdomain interactions, frequently by means of multiple highly conserved residues. The crystal structure also reveals that the active site residue Cys-199 of the KS domain is separated from the active site residue Ser-642 of the AT domain by approximately 80 Angstrom. This distance is too large to be covered simply by alternative positioning of a statically anchored, fully extended phosphopantetheine arm of the acyl carrier protein domain from module 5. Thus, substantial domain reorganization appears necessary for the acyl carrier protein to interact successively with both the AT and the KS domains of this prototypical polyketide synthase module. The 2.7-Angstrom KS-AT structure is fully consistent with a recently reported lower resolution, 4.5-Angstrom model of fatty acid synthase structure, and emphasizes the close biochemical and structural similarity between polyketide synthase and fatty acid synthase enzymology.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures


References
-
- Walsh C. T. Science. 2004;303:1805–1810. - PubMed
-
- Cane D. E., Walsh C. T., Khosla C. Science. 1998;282:63–68. - PubMed
-
- Asturias F. J., Chadick J. Z., Cheung I. K., Stark H., Witkowski A., Joshi A. K., Smith S. Nat. Struct. Mol. Biol. 2005;12:225–232. - PubMed
-
- Cortes J., Haydock S. F., Roberts G. A., Bevitt D. J., Leadlay P. F. Nature. 1990;348:176–178. - PubMed
-
- Donadio S., Staver M. J., McAlpine J. B., Swanson S. J., Katz L. Science. 1991;252:675–679. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

