The Medicago truncatula lysin [corrected] motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes
- PMID: 16844829
- PMCID: PMC1557615
- DOI: 10.1104/pp.106.084657
The Medicago truncatula lysin [corrected] motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes
Erratum in
- Plant Physiol. 2007 Feb;143(2):1078
Abstract
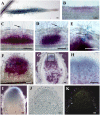
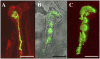
Rhizobial Nod factors are key symbiotic signals responsible for starting the nodulation process in host legume plants. Of the six Medicago truncatula genes controlling a Nod factor signaling pathway, Nod Factor Perception (NFP) was reported as a candidate Nod factor receptor gene. Here, we provide further evidence for this by showing that NFP is a lysin [corrected] motif (LysM)-receptor-like kinase (RLK). NFP was shown both to be expressed in association with infection thread development and to be involved in the infection process. Consistent with deviations from conserved kinase domain sequences, NFP did not show autophosphorylation activity, suggesting that NFP needs to associate with an active kinase or has unusual functional characteristics different from classical kinases. Identification of nine new M. truncatula LysM-RLK genes revealed a larger family than in the nonlegumes Arabidopsis (Arabidopsis thaliana) or rice (Oryza sativa) of at least 17 members that can be divided into three subfamilies. Three LysM domains could be structurally predicted for all M. truncatula LysM-RLK proteins, whereas one subfamily, which includes NFP, was characterized by deviations from conserved kinase sequences. Most of the newly identified genes were found to be expressed in roots and nodules, suggesting this class of receptors may be more extensively involved in nodulation than was previously known.
Figures

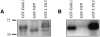
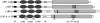


References
-
- Ané JM, Kiss GB, Riely BK, Penmetsa RV, Oldroyd GE, Ayax C, Lévy J, Debellé F, Baek JM, Kalo P, et al (2004) Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 30: 1364–1367 - PubMed
-
- Ardourel M, Demont N, Debelle F, Maillet F, de Billy F, Promé JC, Dénarié J, Truchet G (1994) Rhizobium meliloti lipooligosaccharide nodulation factors: different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses. Plant Cell 6: 1357–1374 - PMC - PubMed
-
- Bateman A, Bycroft M (2000) The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD). J Mol Biol 299: 1113–1119 - PubMed
-
- Ben Amor B, Shaw SL, Oldroyd GE, Maillet F, Penmetsa RV, Cook D, Long SR, Dénarié J, Gough C (2003) The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J 34: 495–506 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

