The multigene family encoding germin-like proteins of barley. Regulation and function in Basal host resistance
- PMID: 16844832
- PMCID: PMC1557593
- DOI: 10.1104/pp.106.083824
The multigene family encoding germin-like proteins of barley. Regulation and function in Basal host resistance
Abstract
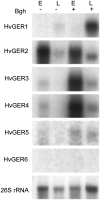
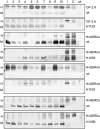
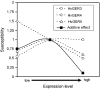
Germin-like proteins (GLPs) have been shown to be encoded by multigene families in several plant species and a role of some subfamily members in defense against pathogen attack has been proposed based on gene regulation studies and transgenic approaches. We studied the function of six GLP subfamilies of barley (Hordeum vulgare) by selecting single mRNAs for gene expression studies as well as overexpression and gene-silencing experiments in barley and Arabidopsis (Arabidopsis thaliana). Expression of all six subfamilies was high in very young seedlings, including roots. The expression pattern gradually changed from developmental to conditional with increasing plant age, whereby pathogen attack and exogenous hydrogen peroxide application were found to be the strongest signals for induction of several GLP subfamilies. Transcripts of four of five GLP subfamilies that are expressed in shoots were predominantly accumulating in the leaf epidermis. Transient overexpression of HvGER4 or HvGER5 as well as transient silencing by RNA interference of HvGER3 or HvGER5 protected barley epidermal cells from attack by the appropriate powdery mildew fungus Blumeria graminis f. sp. hordei. Silencing of HvGER4 induced hypersusceptibility. Transient and stable expression of subfamily members revealed HvGER5 as a new extracellular superoxide dismutase, and protection by overexpression could be demonstrated to be dependent on superoxide dismutase activity of the encoded protein. Data suggest a complex interplay of HvGER proteins in fine regulation of basal resistance against B. graminis.
Figures

Similar articles
-
The germinlike protein GLP4 exhibits superoxide dismutase activity and is an important component of quantitative resistance in wheat and barley.Mol Plant Microbe Interact. 2004 Jan;17(1):109-17. doi: 10.1094/MPMI.2004.17.1.109. Mol Plant Microbe Interact. 2004. PMID: 14714874
-
A class III peroxidase specifically expressed in pathogen-attacked barley epidermis contributes to basal resistance.Mol Plant Pathol. 2008 Sep;9(5):687-96. doi: 10.1111/j.1364-3703.2008.00494.x. Mol Plant Pathol. 2008. PMID: 19018997 Free PMC article.
-
Protein polyubiquitination plays a role in basal host resistance of barley.Plant Cell. 2006 Nov;18(11):3321-31. doi: 10.1105/tpc.106.046326. Epub 2006 Nov 17. Plant Cell. 2006. PMID: 17114351 Free PMC article.
-
Germin-like proteins (GLPs) in cereal genomes: gene clustering and dynamic roles in plant defence.Funct Integr Genomics. 2010 Nov;10(4):463-76. doi: 10.1007/s10142-010-0184-1. Epub 2010 Aug 4. Funct Integr Genomics. 2010. PMID: 20683632 Review.
-
Ramularia collo-cygni--An Emerging Pathogen of Barley Crops.Phytopathology. 2015 Jul;105(7):895-904. doi: 10.1094/PHYTO-11-14-0337-FI. Epub 2015 Jun 29. Phytopathology. 2015. PMID: 25626073 Review.
Cited by
-
Transcriptome-Based Comparative Expression Profiling of Sweet Potato during a Compatible Response with Root-Knot Nematode Meloidogyne incognita Infection.Genes (Basel). 2023 Nov 13;14(11):2074. doi: 10.3390/genes14112074. Genes (Basel). 2023. PMID: 38003017 Free PMC article.
-
Overexpression of Arabidopsis plasmodesmata germin-like proteins disrupts root growth and development.Plant Cell. 2012 Sep;24(9):3630-48. doi: 10.1105/tpc.112.101063. Epub 2012 Sep 7. Plant Cell. 2012. PMID: 22960910 Free PMC article.
-
Transcriptional profiling of Zea mays roots reveals roles for jasmonic acid and terpenoids in resistance against Phytophthora cinnamomi.Funct Integr Genomics. 2013 Jun;13(2):217-28. doi: 10.1007/s10142-013-0314-7. Epub 2013 Feb 22. Funct Integr Genomics. 2013. PMID: 23430324
-
A germin-like protein gene (CchGLP) of Capsicum chinense Jacq. is induced during incompatible interactions and displays Mn-superoxide dismutase activity.Int J Mol Sci. 2011;12(11):7301-13. doi: 10.3390/ijms12117301. Epub 2011 Oct 25. Int J Mol Sci. 2011. PMID: 22174599 Free PMC article.
-
Genome-wide identification of germin-like proteins in peanut (Arachis hypogea L.) and expression analysis under different abiotic stresses.Front Plant Sci. 2023 Jan 23;13:1044144. doi: 10.3389/fpls.2022.1044144. eCollection 2022. Front Plant Sci. 2023. PMID: 36756235 Free PMC article.
References
-
- Baumlein H, Braun H, Kakhovskaya IA, Shutov AD (1995) Seed storage proteins of spermatophytes share a common ancestor with desiccation proteins of fungi. J Mol Evol 41: 1070–1075 - PubMed
-
- Bechtold N, Ellis J, Pelletier G (1993) In-planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. CR Acad Sci Ser III Sci Vie 316: 1194–1199
-
- Berna A, Bernier F (1997) Regulated expression of a wheat germin gene in tobacco: oxalate oxidase activity and apoplastic localization of the heterologous protein. Plant Mol Biol 33: 417–429 - PubMed
-
- Boddu J, Cho S, Kruger WM, Muehlbauer GJ (2006) Transcriptome analysis of the barley-Fusarium graminearum interaction. Mol Plant Microbe Interact 19: 407–417 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

