ZmPIN1a and ZmPIN1b encode two novel putative candidates for polar auxin transport and plant architecture determination of maize
- PMID: 16844839
- PMCID: PMC1557596
- DOI: 10.1104/pp.106.080119
ZmPIN1a and ZmPIN1b encode two novel putative candidates for polar auxin transport and plant architecture determination of maize
Abstract
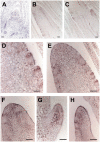
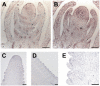
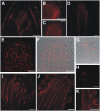
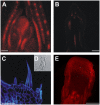
Shoot apical meristems produce organs in a highly stereotypic pattern that involves auxin. Auxin is supposed to be actively transported from cell to cell by influx (AUXIN/LIKE AUXIN proteins) and efflux (PIN-FORMED proteins) membrane carriers. Current hypotheses propose that, at the meristem surface, PIN proteins create patterns of auxin gradients that, in turn, create patterns of gene expression and morphogenesis. These hypotheses are entirely based on work in Arabidopsis (Arabidopsis thaliana). To verify whether these models also apply to other species, we studied the behavior of PIN proteins during maize (Zea mays) development. We identified two novel putative orthologs of AtPIN1 in maize and analyzed their expression pattern during development. The expression studies were complemented by immunolocalization studies using an anti-AtPIN1 antibody. Interestingly, the maize proteins visualized by this antibody are almost exclusively localized in subepidermal meristematic layers. Both tassel and ear were characterized by a compact group of cells, just below the surface, carrying PIN. In contrast to or to complement what was shown in Arabidopsis, these results point to the importance of internally localized cells in the patterning process. We chose the barren inflorescence2 (bif2) maize mutant to study the role of auxin polar fluxes in inflorescence development. In severe alleles of bif2, the tassel and the ear present altered ZmPIN1a and ZmPIN1b protein expression and localization patterns. In particular, the compact groups of cells in the tassel and ear of the mutant were missing. We conclude that BIF2 is important for PIN organization and could play a role in the establishment of polar auxin fluxes in maize inflorescence, indirectly modulating the process of axillary meristem formation and development.
Figures


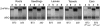

Similar articles
-
BARREN INFLORESCENCE2 interaction with ZmPIN1a suggests a role in auxin transport during maize inflorescence development.Plant Cell Physiol. 2009 Mar;50(3):652-7. doi: 10.1093/pcp/pcp006. Epub 2009 Jan 19. Plant Cell Physiol. 2009. PMID: 19153156
-
barren inflorescence2 Encodes a co-ortholog of the PINOID serine/threonine kinase and is required for organogenesis during inflorescence and vegetative development in maize.Plant Physiol. 2007 Jun;144(2):1000-11. doi: 10.1104/pp.107.098558. Epub 2007 Apr 20. Plant Physiol. 2007. PMID: 17449648 Free PMC article.
-
The Relationship between auxin transport and maize branching.Plant Physiol. 2008 Aug;147(4):1913-23. doi: 10.1104/pp.108.121541. Epub 2008 Jun 11. Plant Physiol. 2008. PMID: 18550681 Free PMC article.
-
The role of PIN auxin efflux carriers in polar auxin transport and accumulation and their effect on shaping maize development.Mol Plant. 2012 Jul;5(4):787-98. doi: 10.1093/mp/ssr103. Epub 2011 Dec 19. Mol Plant. 2012. PMID: 22186966 Review.
-
Auxin: a major regulator of organogenesis.C R Biol. 2010 Apr;333(4):290-6. doi: 10.1016/j.crvi.2010.01.004. Epub 2010 Mar 12. C R Biol. 2010. PMID: 20371103 Review.
Cited by
-
Expression of KxhKN4 and KxhKN5 genes in Kalanchoë blossfeldiana 'Molly' results in novel compact plant phenotypes: towards a cisgenesis alternative to growth retardants.Plant Cell Rep. 2011 Dec;30(12):2267-79. doi: 10.1007/s00299-011-1132-9. Epub 2011 Aug 18. Plant Cell Rep. 2011. PMID: 21850596
-
NPH3- and PGP-like genes are exclusively expressed in the apical tip region essential for blue-light perception and lateral auxin transport in maize coleoptiles.J Exp Bot. 2011 Jun;62(10):3459-66. doi: 10.1093/jxb/err019. Epub 2011 Mar 31. J Exp Bot. 2011. PMID: 21459767 Free PMC article.
-
Enhancing auxin accumulation in maize root tips improves root growth and dwarfs plant height.Plant Biotechnol J. 2018 Jan;16(1):86-99. doi: 10.1111/pbi.12751. Epub 2017 Jun 23. Plant Biotechnol J. 2018. PMID: 28499064 Free PMC article.
-
Maize LAZY1 mediates shoot gravitropism and inflorescence development through regulating auxin transport, auxin signaling, and light response.Plant Physiol. 2013 Nov;163(3):1306-22. doi: 10.1104/pp.113.227314. Epub 2013 Oct 2. Plant Physiol. 2013. PMID: 24089437 Free PMC article.
-
Fluorescent reporter lines for auxin and cytokinin signalling in barley (Hordeum vulgare).PLoS One. 2018 Apr 25;13(4):e0196086. doi: 10.1371/journal.pone.0196086. eCollection 2018. PLoS One. 2018. PMID: 29694399 Free PMC article.
References
-
- Benkova E, Michniewicz M, Sauer M, Teichmann T, Seifertova D, Jurgens G, Friml J (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 - PubMed
-
- Bennett MJ, Marchant A, Green HG, May ST, Ward SP, Millner PA, Walker AR, Schulz B, Feldmann KA (1996) Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 273: 948–950 - PubMed
-
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B (2005) The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 - PubMed
-
- Boutté Y, Crosnier MT, Carraro N, Traas J, Satiat-Jeunemaitre B (2006) The plasma membrane recycling pathway and cell polarity in plants: studies on PIN proteins. J Cell Sci 119: 1255–1265 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

