Functional analyses of two tomato APETALA3 genes demonstrate diversification in their roles in regulating floral development
- PMID: 16844904
- PMCID: PMC1533988
- DOI: 10.1105/tpc.106.042978
Functional analyses of two tomato APETALA3 genes demonstrate diversification in their roles in regulating floral development
Abstract
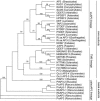
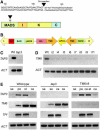
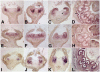
The floral homeotic APETALA3 (AP3) gene in Arabidopsis thaliana encodes a MADS box transcription factor required for specifying petal and stamen identities. AP3 is a member of the euAP3 lineage, which arose by gene duplication coincident with radiation of the core eudicots. Although Arabidopsis lacks genes in the paralogous Tomato MADS box gene 6 (TM6) lineage, tomato (Solanum lycopersicum) possesses both euAP3 and TM6 genes, which have functionally diversified. A loss-of-function mutation in Tomato AP3 (TAP3) resulted in homeotic transformations of both petals and stamens, whereas RNA interference-induced reduction in TM6 function resulted in flowers with homeotic defects primarily in stamens. The functional differences between these genes can be ascribed partly to different expression domains. When overexpressed in an equivalent domain, both genes can partially rescue the tap3 mutant, indicating that relative levels as well as spatial patterns of expression contribute to functional differences. Our results also indicate that the two proteins have differing biochemical capabilities. Together, these results suggest that TM6 and TAP3 play qualitatively different roles in floral development; they also support the ideas that the ancestral role of AP3 lineage genes was in specifying stamen development and that duplication and divergence in the AP3 lineage allowed for the acquisition of a role in petal specification in the core eudicots.
Figures


References
-
- Ambrose, B.A., Lerner, D.R., Ciceri, P., Padilla, C.M., Yanofsky, M.F., and Schmidt, R.J. (2000). Molecular and genetic analyses of the Silky1 gene reveal conservation in floral organ specification between eudicots and monocots. Mol. Cell 5 569–579. - PubMed
-
- Benfey, P.N., and Chua, N.-H. (1990). The cauliflower mosaic virus 35S promoter: Combinatorial regulation of transcription in plants. Science 250 959–966. - PubMed
-
- Brukhin, V., Hernould, M., Gonzalez, N., Chevalier, C., and Mouras, A. (2003). Flower development schedule in tomato Lycopersicon esculentum cv. Sweet Cherry. Sex. Plant Reprod. 15 311–320.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

