Analysis of the Petunia TM6 MADS box gene reveals functional divergence within the DEF/AP3 lineage
- PMID: 16844905
- PMCID: PMC1533978
- DOI: 10.1105/tpc.106.042937
Analysis of the Petunia TM6 MADS box gene reveals functional divergence within the DEF/AP3 lineage
Abstract
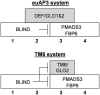
Antirrhinum majus DEFICIENS (DEF) and Arabidopsis thaliana APETALA3 (AP3) MADS box proteins are required to specify petal and stamen identity. Sampling of DEF/AP3 homologs revealed two types of DEF/AP3 proteins, euAP3 and TOMATO MADS BOX GENE6 (TM6), within core eudicots, and we show functional divergence in Petunia hybrida euAP3 and TM6 proteins. Petunia DEF (also known as GREEN PETALS [GP]) is expressed mainly in whorls 2 and 3, and its expression pattern remains unchanged in a blind (bl) mutant background, in which the cadastral C-repression function in the perianth is impaired. Petunia TM6 functions as a B-class organ identity protein only in the determination of stamen identity. Atypically, Petunia TM6 is regulated like a C-class rather than a B-class gene, is expressed mainly in whorls 3 and 4, and is repressed by BL in the perianth, thereby preventing involvement in petal development. A promoter comparison between DEF and TM6 indicates an important change in regulatory elements during or after the duplication that resulted in euAP3- and TM6-type genes. Surprisingly, although TM6 normally is not involved in petal development, 35S-driven TM6 expression can restore petal development in a def (gp) mutant background. Finally, we isolated both euAP3 and TM6 genes from seven solanaceous species, suggesting that a dual euAP3/TM6 B-function system might be the rule in the Solanaceae.
Figures





References
-
- Ambrose, B.A., Lerner, D.R., Ciceri, P., Padilla, C.M., Yanofsky, M.F., and Schmidt, R.J. (2000). Molecular and genetic analyses of the silky1 gene reveal conservation in floral organ specification between eudicots and monocots. Mol. Cell 5 569–579. - PubMed
-
- Bowman, J.L., Sakai, H., Jack, T., Weigel, D., Mayer, U., and Meyerowitz, E.M. (1992). SUPERMAN, a regulator of floral homeotic genes in Arabidopsis. Development 114 599–615. - PubMed
-
- Bradley, D., Carpenter, R., Sommer, H., Hartley, N., and Coen, E. (1993). Complementary floral homeotic phenotypes result from opposite orientations of a transposon at the plena locus of Antirrhinum. Cell 72 85–95. - PubMed
-
- Causier, B., Castillo, R., Zhou, J., Ingram, R., Xue, Y., Schwarz-Sommer, Z., and Davies, B. (2005). Evolution in action: Following function in duplicated floral homeotic genes. Curr. Biol. 15 1508–1512. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

