3dSS: 3D structural superposition
- PMID: 16844975
- PMCID: PMC1538824
- DOI: 10.1093/nar/gkl036
3dSS: 3D structural superposition
Abstract
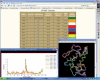
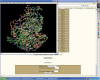
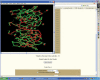
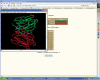
3dSS is a web-based interactive computing server, primarily designed to aid researchers, to superpose two or several 3D protein structures. In addition, the server can be effectively used to find the invariant and common water molecules present in the superposed homologous protein structures. The molecular visualization tool RASMOL is interfaced with the server to visualize the superposed 3D structures with the water molecules (invariant or common) in the client machine. Furthermore, an option is provided to save the superposed 3D atomic coordinates in the client machine. To perform the above, users need to enter Protein Data Bank (PDB)-id(s) or upload the atomic coordinates in PDB format. This server uses a locally maintained PDB anonymous FTP server that is being updated weekly. This program can be accessed through our Bioinformatics web server at the URL http://cluster.physics.iisc.ernet.in/3dss/ or http://10.188.1.15/3dss/.
Figures
Similar articles
-
BSDD: Biomolecules Segment Display Device--a web-based interactive display tool.Nucleic Acids Res. 2004 Jul 1;32(Web Server issue):W645-8. doi: 10.1093/nar/gkh420. Nucleic Acids Res. 2004. PMID: 15215468 Free PMC article.
-
SuperPose: a simple server for sophisticated structural superposition.Nucleic Acids Res. 2004 Jul 1;32(Web Server issue):W590-4. doi: 10.1093/nar/gkh477. Nucleic Acids Res. 2004. PMID: 15215457 Free PMC article.
-
SEM (Symmetry Equivalent Molecules): A web-based GUI to generate and visualize the macromolecules.Nucleic Acids Res. 2003 Jul 1;31(13):3356-8. doi: 10.1093/nar/gkg587. Nucleic Acids Res. 2003. PMID: 12824326 Free PMC article.
-
Seeing the PDB.J Biol Chem. 2021 Jan-Jun;296:100742. doi: 10.1016/j.jbc.2021.100742. Epub 2021 May 4. J Biol Chem. 2021. PMID: 33957126 Free PMC article. Review.
-
QSalignWeb: A Server to Predict and Analyze Protein Quaternary Structure.Front Mol Biosci. 2022 Jan 5;8:787510. doi: 10.3389/fmolb.2021.787510. eCollection 2021. Front Mol Biosci. 2022. PMID: 35071324 Free PMC article. Review.
Cited by
-
Bioinformatics Approaches Applied to the Discovery of Antifungal Peptides.Antibiotics (Basel). 2023 Mar 13;12(3):566. doi: 10.3390/antibiotics12030566. Antibiotics (Basel). 2023. PMID: 36978434 Free PMC article.
-
Evolution, homology conservation, and identification of unique sequence signatures in GH19 family chitinases.J Mol Evol. 2010 May;70(5):466-78. doi: 10.1007/s00239-010-9345-z. Epub 2010 May 18. J Mol Evol. 2010. PMID: 20480157
-
Discovery of novel inhibitors for Nek6 protein through homology model assisted structure based virtual screening and molecular docking approaches.ScientificWorldJournal. 2014 Jan 22;2014:967873. doi: 10.1155/2014/967873. eCollection 2014. ScientificWorldJournal. 2014. PMID: 24587765 Free PMC article.
-
Evaluation of an antimicrobial L-amino acid oxidase and peptide derivatives from Bothropoides mattogrosensis pitviper venom.PLoS One. 2012;7(3):e33639. doi: 10.1371/journal.pone.0033639. Epub 2012 Mar 16. PLoS One. 2012. PMID: 22438972 Free PMC article.
-
The 1.4 A resolution structure of Paracoccus pantotrophus pseudoazurin.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010 Jun 1;66(Pt 6):627-35. doi: 10.1107/S1744309110013989. Epub 2010 May 25. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010. PMID: 20516588 Free PMC article.
References
-
- Bernstein F.C., Koetzle T.F., Williams G.J.B., Meyer E.F., Jr, Brice M.D., Rogers J.R., Kennard O., Shimanouchi T., Tasumi M.J. The Protein Data Bank: a computer based archival file for macromolecular structures. J. Mol. Biol. 1977;112:535–542. - PubMed
-
- Kabsch W.A. Discussion of solution for best rotation of two vectors. Acta Crystallogr. 1978;A34:827–828.
-
- MacLachlan A.D. Rapid comparison of protein structures. Acta Crystallogr. 1982;A38:871–873.
-
- Kearsley S.K. An algorithm for the simultaneous superposition of a structural series. J. Comput. Chem. 1990;11:1187–1192.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

